Detection of Circulating Serum Protein Biomarkers of Non-Muscle Invasive Bladder Cancer after Protein Corona-Silver Nanoparticles Analysis by SWATH-MS
Abstract
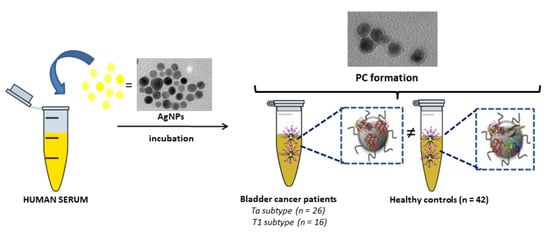
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Biological Samples
2.3. Synthesis of Citrate-Silver Nanoparticles (AgNPs, 9.73 ± 1.70 nm) and Ex Vivo Protein Corona Formation
2.4. Quantification of the Proteins Presented in the Corona-Coated AgNPs by SWATH-MS
2.5. Protein Functional Interaction Network Analysis
3. Results and Discussion
3.1. Incubation of AgNPs (9.73 ± 1.70 nm) with HS Samples: Ex Vivo Protein Corona Formation and Characterization
3.2. Quantitative Analysis of the Protein Corona-Coated AgNPs by SWATH-MS
3.3. The Biological Role of the NMIBC-Related Proteins Identified in the AgNPs-Protein Corona
- (a)
- complements: C1r subcomponent (C1R), complement C1s subcomponent (C1S), complement C2 (C2), complement C4-A (C4A), complement C4-B (C4B), C4b-binding protein alpha chain (C4BPA), complement C5 (C5), complement component C6 (C6), complement component C7 (C7), complement component C8 beta chain (C8B), complement component C8 gamma chain (C8G), complement component C9 (C9), complement factor B (CFB), complement factor H (CFH), and complement factor I (CFI);
- (b)
- coagulation factors: coagulation factor II (F2), coagulation factor IX (F9) coagulation factor (F10) and coagulation factor XII (F12);
- (c)
- serine protease related proteins: alpha-1-antitrypsin (SERPINA1), antithrombin-III (SERPINC1), alpha-2-antiplasmin (SERPINF2), plasma protease C1 inhibitor (SERPING1), plasma kallikrein (KLKB1) and plasminogen (PLG);
- (d)
- vitamin K-dependent proteins: vitamin K-dependent protein C (PROC) and vitamin K-dependent protein S (PROS1); and
- (e)
- glycoproteins: fibrinogen alpha chain (FGA) and vitronectin (VTN).
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [Green Version]
- Aldousari, S.; Kassouf, W. Update on the management of non-muscle invasive bladder cancer. Can. Urol. Assoc. J. 2010, 4, 56–64. [Google Scholar] [CrossRef] [Green Version]
- Chamie, K.; Litwin, M.S.; Bassett, J.C.; Daskivich, T.J.; Lai, J.; Hanley, J.M.; Konety, B.R.; Saigal, C.S. Urologic Diseases in America Project. Recurrence of high-risk bladder cancer: A population-based analysis. Cancer 2013, 119, 3219–3227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, W.S.; Rodney, S.; Lamb, B.; Feneley, M.; Kelly, J. Management of non-muscle invasive bladder cancer: A comprehensive analysis of guidelines from the United States, Europe and Asia. Cancer Treat. Rev. 2016, 47, 22–31. [Google Scholar] [CrossRef]
- Svatek, R.S.; Hollenbeck, B.K.; Holmäng, S.; Lee, R.; Kim, S.P.; Stenzl, A.; Lotan, Y. The economics of bladder cancer: Costs and considerations of caring for this disease. Eur. Urol. 2014, 66, 253–262. [Google Scholar] [CrossRef]
- Hanash, S.M.; Pitteri, S.J.; Faca, V.M. Mining the plasma proteome for cancer biomarkers. Nature 2008, 452, 571–579. [Google Scholar] [CrossRef] [PubMed]
- Wulfuhle, J.D.; Liotta, L.A.; Petricoin, E.F. Proteomic applications for the early detection of cancer. Nat. Rev. Cancer 2003, 3, 267–275. [Google Scholar] [CrossRef]
- Tadikkaran, L.; Siegenthaler, M.A.; Crettaz, D.; Queloz, P.-A.; Schneider, P.; Tissot, J.-D. Recent advances in blood-related proteomics. Proteomics 2005, 5, 3019–3034. [Google Scholar] [CrossRef] [PubMed]
- Buszewska-Forajta, M.; Pomastowski, P.; Monedeiro, F.; Walczak-Skierska, J.; Markuszewski, M.; Matuszewski, M.; Markuszewski, M.J.; Buszewski, B. Lipidomics as a Diagnostic Tool for Prostate Cancer. Cancers 2021, 13, 2000. [Google Scholar] [CrossRef]
- Kitamura, Y.; Usami, R.; Ichihara, S.; Kida, H.; Satoh, M.; Tomimoto, H.; Murata, M.; Oikawa, S. Plasma protein profling for potential biomarkers in the early diagnosis of Alzheimer’s disease. Neurol. Res. 2017, 39, 231–238. [Google Scholar] [CrossRef]
- Barrachina, M.N.; Sueiro, A.M.; Casas, V.; Izquierdo, I.; Hermida-Nogueira, L.; Guitián, E.; Casanueva, F.F.; Abián, J.; Carrascal, M.; Pardo, M.; et al. A combination of proteomic approaches identifes a panel of circulating extracellular vesicle proteins related to the risk of sufering cardiovascular disease in obese patients. Proteomics 2019, 19, e1800248. [Google Scholar] [PubMed] [Green Version]
- Tan, G.; Tan, G.; Wang, H.; Yuan, J.; Qin, W.; Dong, X.; Wu, H.; Meng, P. Tree serum metabolite signatures for diagnosing low-grade and high-grade bladder cancer. Sci. Rep. 2017, 7, 46176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frantzi, M.; Vlahou, A. Ten years of proteomics in bladder cancer: Progress and future directions. Bladder Cancer 2017, 3, 1–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, L.; Huang, Z.; Gao, Y.; Chen, Y.; Hang, W.; Xing, J.; Yan, X. LC-MS-based serum metabolic profiling for genitourinary cancer classification and cancer type-specific biomarker discovery. Proteomics 2012, 12, 2238–2246. [Google Scholar] [CrossRef]
- Nedjadi, T.; Benabdelkamal, H.; Albarakati, N.; Masood, A.; Al-Sayyad, A.; Alfadda, A.A.; Alanazi, I.O.; Al-Ammari, A.; Al-Maghrabi, J. Circulating proteomic signature for detection of biomarkers in bladder cancer patients. Sci. Rep. 2020, 10, 10999. [Google Scholar] [CrossRef]
- Luo, Y.; Mok, T.S.; Lin, X.; Zhang, W.; Cui, Y.; Guo, J.; Chen, X.; Zhang, T.; Wang, T. SWATH-based proteomics identified carbonic anhydrase 2 as a potential diagnosis biomarker for nasopharyngeal carcinoma. Sci. Rep. 2017, 7, 41191. [Google Scholar] [CrossRef]
- Diamandis, E.P. Mass spectrometry as a diagnostic and a cancer biomarker discovery tool opportunities and potential limitations. Mol. Cell Proteom. 2004, 3, 367–378. [Google Scholar] [CrossRef] [Green Version]
- Ralhan, R.; DeSouza, L.V.; Matta, A.; Tripathi, S.C.; Ghanny, S.; Gupta, S.D.; Bahadur, S.; Siu, K.M. Discovery and verification of head-and-neck cancer biomarkers by differential protein expression analysis using iTRAQ labeling, multidimensional liquid chromatography, and tandem mass spectrometry. Mol. Cell Proteom. 2008, 7, 1162–1173. [Google Scholar] [CrossRef] [Green Version]
- Ludwig, C.; Gillet, L.; Rosenberger, G.; Amon, S.; Collins, B.C.; Aebersold, R. Data-independent acquisition-based SWATH-MS for quantitative proteomics: A tutorial. Mol. Syst. Biol. 2018, 14, e8126. [Google Scholar] [CrossRef]
- Meyer, J.G.; Schilling, B. Clinical applications of quantitative proteomics using targeted and untargeted data-independent acquisition techniques. Expert Rev. Proteom. 2017, 14, 419–429. [Google Scholar] [CrossRef]
- Narasimhan, M.; Kannan, S.; Chawade, A.; Bhattacharjee, A.; Govekar, R. Clinical biomarker discovery by SWATH-MS based label-free quantitative proteomics: Impact of criteria for identification of differentiators and data normalization method. J. Transl. Med. 2019, 17, 184. [Google Scholar] [CrossRef] [PubMed]
- Rifai, N.; Gillette, M.A.; Carr, S.A. Protein biomarker discovery and validation: The long and uncertain path to clinical utility. Nat. Biotechnol. 2006, 24, 971–983. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Lu, H.; Yang, P. Recent developments of nanoparticle-based enrichment methods for mass spectrometric analysis in proteomic. Sci. China Chem. 2010, 53, 695–703. [Google Scholar] [CrossRef]
- Docter, D.; Westmeier, D.; Markiewicz, M.; Stolte, S.; Knauer, S.K.; Stauber, R.H. The nanoparticle biomolecule corona: Lessons learned—Challenge accepted? Chem. Soc. Rev. 2015, 44, 6094–6121. [Google Scholar] [CrossRef] [Green Version]
- Corbo, C.; Molinaro, R.; Tabatabaei, M.; Farokhzad, O.C.; Mahmoudi, M. Personalized protein corona on nanoparticles and its clinical implications. Biomater. Sci. 2017, 5, 378–387. [Google Scholar] [CrossRef] [PubMed]
- Chantada-Vázquez, M.P.; Castro-López, A.; Bravo, S.B.; Vázquez-Estévez, S.; Acea-Nebril, B.; Núñez, C. Proteomic analysis of the bio-corona formed on the surface of (Au, Ag, Pt)-nanoparticles in human serum. Colloids Surf. B 2019, 177, 141–148. [Google Scholar] [CrossRef] [PubMed]
- Chantada-Vázquez, M.P.; Castro-López, A.; García-Vence, M.; Vázquez-Estévez, S.; Acea-Nebril, B.; Calatayud, D.G.; Jardiel, T.; Bravo, S.B.; Núñez, C. Proteomic investigation on bio-corona of Au, Ag and Fe nanoparticles for the discovery of triple negative breast cancer serum protein biomarkers. J. Proteom. 2020, 212, 103581. [Google Scholar] [CrossRef]
- Chantada-Vázquez, M.P.; García-Vence, M.; Vázquez-Estévez, S.; Bravo, S.B.; Núñez, C. Identification of a Profile of Neutrophil-Derived Granule Proteins in the Surface of Gold Nanoparticles after Their Interaction with Human Breast Cancer Sera. Nanomaterials 2020, 10, 1223. [Google Scholar] [CrossRef]
- Chantada-Vázquez, M.P.; Castro-López, A.; García-Vence, M.; Acea-Nebril, B.; Bravo, S.B.; Núñez, C. Protein Corona Gold Nanoparticles Fingerprinting Reveals a Profile of Blood Coagulation Proteins in the Serum of HER2-Overexpressing Breast Cancer Patients. Int. J. Mol. Sci. 2020, 21, 8449. [Google Scholar] [CrossRef]
- García-Vence, M.; Chantada-Vázquez, M.P.; Cameselle-Teijeiro, J.M.; Bravo, S.B.; Núñez, C. A novel nanoproteomic approach for the identification of molecular targets associated with thyroid tumors. Nanomaterials 2020, 10, 2370. [Google Scholar] [CrossRef]
- Bastus, N.G.; Merkoci, F.; Piella, J.; Puntes, V. Synthesis of highly monodisperse citrate- stabilized silver nanoparticles of up to 200 nm: Kinetic control and catalytic properties. Chem. Mater. 2014, 26, 2836–2846. [Google Scholar] [CrossRef]
- Papafilippou, L.; Claxton, A.; Dark, P.; Kostarelos, K.; Hadjidemetriou, M. Protein corona fingerprinting to differentiate sepsis from non-infectious systemic inflammation. Nanoscale 2020, 12, 10240–10253. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef] [PubMed]
- Dobrovolskaia, M.A.; Patri, A.K.; Zheng, J.; Clogston, J.D.; Ayub, N.; Aggarwal, P.; Neun, B.W.; Hall, J.B.; McNeil, S.E. Interaction of colloidal gold nanoparticles with human blood: Effects on particle size and analysis of plasma protein binding profiles. Nanomedicine 2009, 5, 106–117. [Google Scholar] [CrossRef] [Green Version]
- García-Álvarez, R.; Hadjidemetriou, M.; Sánchez-Iglesias, A.; Liz-Marzán, L.M.; Kostarelos, K. In vivo formation of protein corona on gold nanoparticles: The effect of size and shape. Nanoscale 2018, 10, 1256–1264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krem, M.M.; Di Cera, E. Evolution of enzyme cascades from embryonic development to blood coagulation. Trends Biochem. Sci. 2002, 27, 67–74. [Google Scholar] [CrossRef]
- Sjöberg, A.P.; Trouw, L.A.; Blom, A.M. Complement activation and inhibition: A delicate balance. Trends Immunol. 2009, 30, 83–90. [Google Scholar] [CrossRef]
- Zipfel, P.F.; Skerka, C. Complement regulators and inhibitory proteins. Nat. Rev. Immunol. 2009, 9, 729–740. [Google Scholar] [CrossRef]
- Afshar-Kharghan, V. The role of the complement system in cancer. J. Clin. Investig. 2017, 127, 780–789. [Google Scholar] [CrossRef] [Green Version]
- Pio, R.; Ajona, D.; Lambris, J.D. Complement inhibition in cancer therapy. Semin. Immunol. 2013, 25, 54–64. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.-L.; Lin, T.-S.; Tsai, C.-H.; Wu, C.-C.; Chung, T.; Chien, K.-Y.; Wu, M.; Chang, Y.-S.; Yu, J.-S.; Chen, Y.-T. Identification of potential bladder cancer markers in urine by abundant-protein depletion coupled with quantitative proteomics. J. Proteom. 2013, 85, 28–43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, Z.-Z.; Corey, M.J.; Pärepalo, M.; Majno, S.; Hellwage, J.; Zipfel, P.F.; Kinders, R.J.; Raitanen, M.; Meri, S.; Jokiranta, T.S. Complement Factor H as a Marker for Detection of Bladder Cancer. Clin. Chem. 2005, 51, 856–863. [Google Scholar] [CrossRef] [PubMed]
- Urquidi, V.; Goodison, S.; Ross, S.; Chang, M.; Dai, Y.; Rosser, C.J. Diagnostic Potential of Urinary α1-Antitrypsin and Apolipoprotein E in the Detection of Bladder Cancer. J. Urol. 2012, 188, 2377–2383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lindén, M.; Segersten, U.; Runeson, M.; Wester, K.; Busch, C.; Pettersson, U.; Lind, S.B.; Malmström, P.-U. Tumour expression of bladder cancer-associated urinary proteins. BJU Int. 2013, 112, 407–415. [Google Scholar] [CrossRef]
- Lindén, M.; Lind, S.B.; Mayrhofer, C.; Segersten, U.; Wester, K.; Lyutvinskiy, Y.; Zubarev, R.; Malmström, P.-U.; Pettersson, U. Proteomic analysis of urinary biomarker candidates for non-muscle invasive bladder cancer. Proteomics 2012, 12, 135. [Google Scholar] [CrossRef]
- Ho, Y.Y.; Deckelbaum, R.J.; Chen, Y.; Vogel, T.; Talmag, D.A. Apolipoprotein E inhibits serum-stimulated cell proliferation and enhances serum-independent cell proliferation. J. Biol. Chem. 2001, 276, 43455. [Google Scholar] [CrossRef] [Green Version]
- Mahley, R.W.; Rall, S.C., Jr. Apolipoprotein E: Far more than a lipid transport protein. Annu. Rev. Genom. Hum. Genet. 2000, 1, 507. [Google Scholar] [CrossRef]
- Fredericks, W.J.; McGarvey, T.; Wang, H.; Lal, P.; Puthiyaveettil, R.; Tomaszewski, J.; Sepulveda, J.; Labelle, E.; Weiss, J.S.; Nickerson, M.L.; et al. The bladder tumor suppressor protein TERE1 (UBIAD1) modulates cell cholesterol: Implications for tumor progression. DNA Cell Biol. 2011, 30, 851. [Google Scholar] [CrossRef] [Green Version]
- McGarvey, T.W.; Nguyen, T.B.; Malkowicz, S.B. An interaction between apolipoprotein E and TERE1 with a possible association with bladder tumor formation. J. Cell Biochem. 2005, 95, 419. [Google Scholar] [CrossRef]
- Xue, A.; Chang, J.W.; Chung, L.; Samra, J.; Hugh, T.; Gill, A.; Butturini, G.; Baxter, R.C.; Smith, R.C. Serum apolipoprotein C-II is prognostic for survival after pancreatic resection for adenocarcinoma. Br. J. Cancer 2012, 107, 1883–1891. [Google Scholar] [CrossRef] [Green Version]
- Hachem, H.; Favre, G.; Ghalim, N.; Puchois, P.; Fruchart, J.C.; Soula, G. Quantitative abnormalities of lipoprotein particles in multiple myeloma. J. Clin. Chem. Clin. Biochem. 1987, 25, 675–679. [Google Scholar] [CrossRef]
- Humphries, J.M.; Penno, M.; Weiland, F.; Klingler-Hoffmann, M.; Zuber, A.; Boussioutas, A.; Ernst, M.; Hoffmann, P. Identification and validation of novel candidate protein biomarkers for the detection of human gastric cancer. Biochim. Biophys. Acta 2014, 1844, 1051–1058. [Google Scholar] [CrossRef]
- Honda, K.; Kobayashi, M.; Okusaka, T.; Rinaudo, J.A.; Huang, Y.; Marsh, T.; Sanada, M.; Sasajima, Y.; Nakamori, S.; Shimahara, M.; et al. Plasma biomarker for detection of early stage pancreatic cancer and risk factors for pancreatic malignancy using antibodies for apolipoprotein-aii isoforms. Sci. Rep. 2015, 5, 15921. [Google Scholar] [CrossRef]
- Von Zychlinski, A.; Williams, M.; McCormick, S.; Kleffmann, T. Absolute quantification of apolipoproteins and associated proteins on human plasma lipoproteins. J. Proteom. 2014, 106, 181–190. [Google Scholar] [CrossRef]
- Hu, Y.-W.; Chen, Z.-P.; Hu, X.-M.; Zhao, J.-Y.; Huang, J.-L.; Ma, X.; Li, S.-F.; Qiu, Y.-R.; Wu, X.-J.; Sha, Y.-H.; et al. The miR-573/apoM/Bcl2A1-dependent signal transduction pathway is essential for hepatocyte apoptosis and hepatocarcinogenesis. Apoptosis 2015, 20, 1321–1337. [Google Scholar] [CrossRef]
- Jiang, J.; Wu, C.; Luo, G.; Zheng, L.; Chen, L.; Zhang, X.; Xu, N. Expression of apolipoprotein M in human hepatocellular carcinoma tissues. Acta Histochem. 2011, 113, 53–57. [Google Scholar] [CrossRef] [Green Version]
- Luo, G.; Zhang, X.; Mu, Q.; Chen, L.; Zheng, L.; Wei, J.; Berggren-Söderlund, M.; Nilsson-Ehle, P.; Xu, N. Expression and localization of apolipoprotein m in human colorectal tissues. Lipids Health Dis. 2010, 9, 102. [Google Scholar] [CrossRef] [Green Version]
- Lemańska-Perek, A.; Lis-Kuberka, J.; Lepczyński, A.; Dratwa-Chałupnik, A.; Tupikowski, K.; Kątnik-Prastowska, I.; Ożgo, M. Potential plasma biomarkers of bladder cancer identified by proteomic analysis: A pilot study. Adv. Clin. Exp. Med. 2019, 28, 339–346. [Google Scholar] [CrossRef] [PubMed]
- Mondul, A.M.; Weinstein, S.J.; Virtamo, J.; Albanes, D. Influence of vitamin D binding protein on the association between circulating vitamin D and risk of bladder cancer. Br. J. Cancer 2012, 107, 1589–1594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, D.; Cheng, S.-Q.; Ji, H.-F.; Wang, J.-S.; Xu, H.-T.; Zhang, G.-Q.; Pang, D. Evaluation of protein pigment epithelium-derived factor (PEDF) and microvessel density (MVD) as prognostic indicators in breast cancer. J. Cancer Res. Clin. Oncol. 2010, 136, 1719–1727. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Yao, A.; Zhang, W.; Lu, S.; Yu, Y.; Deng, L.; Yin, A.; Xia, Y.; Sun, B.; Wang, X. Human mesenchymal stem cells overexpressing pigment epithelium-derived factor inhibit hepatocellular cancer in nude mice. Oncogene 2010, 29, 2784–2794. [Google Scholar] [CrossRef] [Green Version]
- Guan, M.; Jiang, H.; Xu, C.; Rong, X.; Zhongqing, C.; Lu, Y. Adenovirus-mediated PEDF expression inhibits prostate cancer cell growth and results in augmented expression of PAI-2. Cancer Biol. Ther. 2007, 6, 419–425. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Chen, S.; Huang, X.; Han, J.; Wang, Q.; Shi, D.; Cheng, R.; Gao, G.; Yang, X. Growth suppression of cervical cancer by pigment epithelium-derived factor via anti-angiogenesis. Cancer Biol. Ther. 2010, 9, 975–977. [Google Scholar] [CrossRef] [Green Version]
- Feng, C.; Ding, Q.; Zhang, Y.; Jiang, H.; Wen, H.; Wang, P.; Wu, Z. Pigment epithelium-derived factor expression is down-regulated in bladder tumors and correlates with vascular endothelial growth factor and matrix metalloproteinase-9. Int. Urol. Nephrol. 2011, 43, 383–390. [Google Scholar] [CrossRef]
- Feng, C.; Wang, P.; Ding, Q.; Guan, M.; Zhang, Y.; Jiang, H.; Wen, H.; Wu, Z. Expression of pigment epithelium-derived factor and tumor necrosis factor-α is correlated in bladder tumor and is related to tumor angiogenesis. Urol. Oncol. 2013, 31, 241–246. [Google Scholar] [CrossRef]
- Badawy, A.M.E.; Luxton, T.P.; Silva, R.G.; Scheckel, K.G.; Suidan, M.T.; Tolaymat, T.M. Impact of Environmental Conditions (pH, Ionic Strength, and Electrolyte Type) on the Surface Charge and Aggregation of Silver Nanoparticles Suspensions. Environ. Sci. Technol. 2010, 44, 1260–1266. [Google Scholar] [CrossRef]
- Axson, J.L.; Stark, D.I.; Bondy, A.L.; Capracotta, S.S.; Maynard, A.D.; Philbert, M.A.; Bergin, I.L.; Ault, A.P. Rapid Kinetics of Size and pH-Dependent Dissolution and Aggregation of Silver Nanoparticles in Simulated Gastric Fluid. J. Phys. Chem. C 2015, 119, 20632–20641. [Google Scholar] [CrossRef] [Green Version]
- Gunsolus, I.L.; Mousavi, M.P.S.; Hussein, K.; Bühlmann, P.; Haynes, C.L. Effects of Humic and Fulvic Acids on Silver Nanoparticle Stability, Dissolution, and Toxicity. Environ. Sci. Technol. 2015, 49, 8078–8086. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, F.; Alvarez, P.J.; Zhu, D. Microbial extracellular polymeric substances reduce Ag+ to silver nanoparticles and antagonize bactericidal activity. Environ. Sci. Technol. 2014, 48, 316–322. [Google Scholar] [CrossRef] [PubMed]
- Fernando, I.; Zhou, Y. Impact of pH on the stability, dissolution and aggregation kinetics of silver nanoparticles. Chemosphere 2019, 216, 297–305. [Google Scholar] [CrossRef] [PubMed]
- Lundqvist, M.; Stigler, J.; Cedervall, T.; Berggård, T.; Flanagan, M.B.; Lynch, I.; Elia, G.; Dawson, K. The evolution of the protein corona around nanoparticles: A test study. ACS Nano 2011, 5, 7503–7509. [Google Scholar] [CrossRef] [PubMed]
- Vroman, L. Effect of adsorbed proteins on the wettability of hydrophilic and hydrophobic solids. Nat. Cell Biol. 1962, 196, 476–477. [Google Scholar] [CrossRef] [PubMed]
- Frost, R.; Langhammer, C.; Cedervall, T. Real-time in situ analysis of biocorona formation and evolution on silica nanoparticles in defined and complex biological environments. Nanoscale 2017, 9, 3620–3628. [Google Scholar] [CrossRef] [Green Version]
- Baimanov, D.; Cai, R.; Chen, C. Understanding the chemical nature of nanoparticle-protein interactions. Bioconjug. Chem. 2019, 30, 1923–1937. [Google Scholar] [CrossRef]
- Kihara, S.; van der Heijden, N.J.; Seal, C.K.; Mata, J.P.; Whitten, A.E.; Köper, I.; McGillivray, D.J. Soft and hard interactions between polystyrene nanoplastics and human serum albumin protein corona. Bioconjug. Chem. 2019, 30, 1067–1076. [Google Scholar] [CrossRef]
- Winzen, S.; Schoettler, S.; Baier, G.; Rosenauer, C.; Mailaender, V.; Landfestera, K.; Mohr, K. Complementary analysis of the hard and soft protein corona: Sample preparation critically effects corona composition. Nanoscale 2015, 7, 2992–3001. [Google Scholar] [CrossRef] [Green Version]
- Park, S.J. Protein—Nanoparticle Interaction: Corona Formation and Conformational Changes in Proteins on Nanoparticles. Int. J. Nanomed. 2020, 15, 5783–5802. [Google Scholar] [CrossRef]
- Johnston, B.D.; Kreyling, W.G.; Pfeiffer, C.; Schäffler, M.; Sarioglu, H.; Ristig, S.; Hirn, S.; Haberl, N.; Thalhammer, S.; Hauck, S.M.; et al. Colloidal stability and surface chemistry are key factors for the composition of the protein corona of inorganic gold nanoparticles. Adv. Funct. Mater. 2017, 27, 1701956. [Google Scholar] [CrossRef]
- Nienhaus, K.; Nienhaus, G.U. Towards a molecular-level understanding of the protein corona around nanoparticles—Recent advances and persisting challenges. Curr. Opin. Biomed. Eng. 2019, 10, 11–22. [Google Scholar] [CrossRef]
- Walkey, C.D.; Olsen, J.B.; Guo, H.; Emili, A.; Chan, W.C. Nanoparticle size and surface chemistry determine serum protein adsorption and macrophage uptake. J. Am. Chem. Soc. 2012, 134, 2139–2147. [Google Scholar] [CrossRef]
- Su, G.; Zhou, X.; Zhou, H.; Li, Y.; Zhang, X.; Liu, Y.; Cao, D.; Yan, B. Size-dependent facilitation of cancer cell targeting by proteins adsorbed on nanoparticles. ACS Appl. Mater. Interfaces 2016, 8, 30037–30047. [Google Scholar] [CrossRef] [PubMed]
- Ashby, J.; Pan, S.; Zhong, W. Size and surface functionalization of iron oxide nanoparticles influence the composition and dynamic nature of their protein corona. ACS Appl. Mater. Interfaces 2014, 6, 15412–15419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vasti, C.; Bedoya, D.A.; Rojas, R.; Giacomelli, C.E. Effect of the protein corona on the colloidal stability and reactivity of LDH-based nanocarriers. J. Mater. Chem. B 2016, 4, 2008–2016. [Google Scholar] [CrossRef]
- Pareek, V.; Bhargava, A.; Bhanot, V.; Gupta, R.; Jain, N.; Panwar, J. Formation and characterization of protein corona around nanoparticles: A review. J. Nanosci. Nanotechnol. 2018, 18, 6653–6670. [Google Scholar] [CrossRef] [PubMed]
- Monopoli, M.P.; Walczyk, D.; Campbell, A.; Elia, G.; Lynch, I.; Bombelli, F.B.; Dawson, K.A. Physical−chemical aspects of protein corona: Relevance to in vitro and in vivo biological impacts of nanoparticles. J. Am. Chem. Soc. 2011, 133, 2525–2534. [Google Scholar] [CrossRef]
- Khan, S.; Gupta, A.; Nandi, C.K. Controlling the fate of protein corona by tuning surface properties of nanoparticles. J. Phys. Chem. Lett. 2013, 4, 3747–3752. [Google Scholar] [CrossRef]
- Kharazian, B.; Hadipour, N.; Ejtehadi, M. Understanding the nanoparticle–protein corona complexes using computational and experimental methods. Int. J. Biochem. Cell Biol. 2016, 75, 162–174. [Google Scholar] [CrossRef]







| SWATH-MS Analysis | ||||||
|---|---|---|---|---|---|---|
| Comparison | Protein Number (p-Value ≤ 0.05) | |||||
| Total | Up-Regulated | Down-Regulated | Specific | Up-Regulated | Down-Regulated | |
| Controls vs. T1 | 40 | 11 | 29 | 16 | 1 | 15 |
| Controls vs. Ta | 46 | 30 | 16 | 22 | 20 | 2 |
| Protein Name | UniProt Name | Gene | p-Value | Fold Change | NMIBC Sybtype |
|---|---|---|---|---|---|
| Beta-2-glycoprotein 1 | APOH_HUMAN | APOH | 0.031038566 | 1.413139568 | ↑ T1 |
| Complement C4-A | CO4A_HUMAN | C4A | 3.35E-05 | 1.872021358 | ↓ T1 |
| Apolipoprotein F | APOF_HUMAN | APOF | 0.000241559 | 2.110329795 | ↓ T1 |
| Complement factor B | CFAB_HUMAN | CFB | 0.000636998 | 1.372510381 | ↓ T1 |
| Complement C4-B | CO4B_HUMAN | C4B | 0.001425038 | 1.622969299 | ↓ T1 |
| Complement component C8 gamma chain | CO8G_HUMAN | C8G | 0.003262992 | 1.467525612 | ↓ T1 |
| Ficolin-2 | FCN2_HUMAN | FCN2 | 0.003600193 | 1.765148338 | ↓ T1 |
| Apolipoprotein E | APOE_HUMAN | APOE | 0.004492395 | 1.342803692 | ↓ T1 |
| Apolipoprotein A-IV | APOA4_HUMAN | APOA4 | 0.004832934 | 1.330273828 | ↓ T1 |
| Hyaluronan-binding protein 2 | HABP2_HUMAN | HABP2 | 0.005064575 | 1.385807696 | ↓ T1 |
| Platelet basic protein | CXCL7_HUMAN | PPBP | 0.007192858 | 1.607064055 | ↓ T1 |
| Pigment epithelium-derived factor | PEDF_HUMAN | SERPINF1 | 0.0177288 | 2.054793133 | ↓ T1 |
| Complement C5 | CO5_HUMAN | C5 | 0.022678206 | 1.261047319 | ↓ T1 |
| Vitamin K-dependent protein C | PROC_HUMAN | PROC | 0.023388014 | 1.233676674 | ↓ T1 |
| Coagulation factor X | FA10_HUMAN | F10 | 0.028581041 | 1.261414143 | ↓ T1 |
| Apolipoprotein A-II | APOA2_HUMAN | APOA2 | 0.029458523 | 2.388772729 | ↓ T1 |
| Fibrinogen alpha chain | FIBA_HUMAN | FGA | 0.00018616 | 1.692142233 | ↑ Ta |
| Complement component C7 | CO7_HUMAN | C7 | 0.0014036 | 1,37740437 | ↑ Ta |
| Vitronectin | VTNC_HUMAN | VTN | 0.00148246 | 1.330829684 | ↑ Ta |
| Complement component C9 | CO9_HUMAN | C9 | 0.00259827 | 1.326995622 | ↑ Ta |
| Vitamin D-binding protein | VTDB_HUMAN | GC | 0.003928 | 1.340366003 | ↑ Ta |
| Complement component C6 | CO6_HUMAN | C6 | 0.0042717 | 1.446886498 | ↑ Ta |
| Complement component C8 beta chain | CO8B_HUMAN | C8B | 0.00538557 | 1.255996294 | ↑ Ta |
| Hemoglobin subunit alpha | HBA_HUMAN | HBA1 | 0.00590282 | 5.020407249 | ↑ Ta |
| Galectin-3-binding protein | LG3BP_HUMAN | LGALS3BP | 0.00753614 | 2.016590504 | ↑ Ta |
| Inter-alpha-trypsin inhibitor heavy chain H1 | ITIH1_HUMAN | ITIH1 | 0.00788349 | 0.719608487 | ↑ Ta |
| Complement factor I | CFAI_HUMAN | CFI | 0.00810041 | 1.222227898 | ↑ Ta |
| Immunoglobulin lambda variable 3–27 | LV327_HUMAN | IGLV3-27 | 0.01005932 | 1.783180848 | ↑ Ta |
| Hemoglobin subunit beta | HBB_HUMAN | HBB | 0.01030231 | 4.673494773 | ↑ Ta |
| Alpha-1-antichymotrypsin | AACT_HUMAN | SERPINA3 | 0.01546691 | 5.505121842 | ↑ Ta |
| C4b-binding protein alpha chain | C4BPA_HUMAN | C4BPA | 0.01766023 | 1.267495989 | ↑ Ta |
| Alpha-1B-glycoprotein | A1BG_HUMAN | A1BG | 0.03046776 | 1.640307834 | ↑ Ta |
| Alpha-1-antitrypsin | A1AT_HUMAN | SERPINA1 | 0.03633694 | 3.239474829 | ↑ Ta |
| Ceruloplasmin | CERU_HUMAN | CP | 0.03816982 | 1.652775639 | ↑ Ta |
| Immunoglobulin heavy variable 3-30-5 | HV335_HUMAN | IGHV3-35 | 0.04091296 | 1.749806744 | ↑ Ta |
| Complement factor H | CFAH_HUMAN | CFH | 0.04493203 | 1.213794163 | ↑ Ta |
| Vitamin K-dependent protein S | PROS_HUMAN | PROS1 | 0.00653335 | 1.241725216 | ↓ Ta |
| Alpha-2-antiplasmin | A2AP_HUMAN | SERPINF2 | 0.00979701 | 1.476811908 | ↓ Ta |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gómez, B.B.; López-Cortés, R.; Casas-Nebra, F.J.; Vázquez-Estévez, S.; Pérez-Fentes, D.; Chantada-Vázquez, M.d.P.; Bravo, S.B.; Núñez, C. Detection of Circulating Serum Protein Biomarkers of Non-Muscle Invasive Bladder Cancer after Protein Corona-Silver Nanoparticles Analysis by SWATH-MS. Nanomaterials 2021, 11, 2384. https://doi.org/10.3390/nano11092384
Gómez BB, López-Cortés R, Casas-Nebra FJ, Vázquez-Estévez S, Pérez-Fentes D, Chantada-Vázquez MdP, Bravo SB, Núñez C. Detection of Circulating Serum Protein Biomarkers of Non-Muscle Invasive Bladder Cancer after Protein Corona-Silver Nanoparticles Analysis by SWATH-MS. Nanomaterials. 2021; 11(9):2384. https://doi.org/10.3390/nano11092384
Chicago/Turabian StyleGómez, Benito Blanco, Rubén López-Cortés, Francisco Javier Casas-Nebra, Sergio Vázquez-Estévez, Daniel Pérez-Fentes, María del Pilar Chantada-Vázquez, Susana B. Bravo, and Cristina Núñez. 2021. "Detection of Circulating Serum Protein Biomarkers of Non-Muscle Invasive Bladder Cancer after Protein Corona-Silver Nanoparticles Analysis by SWATH-MS" Nanomaterials 11, no. 9: 2384. https://doi.org/10.3390/nano11092384
APA StyleGómez, B. B., López-Cortés, R., Casas-Nebra, F. J., Vázquez-Estévez, S., Pérez-Fentes, D., Chantada-Vázquez, M. d. P., Bravo, S. B., & Núñez, C. (2021). Detection of Circulating Serum Protein Biomarkers of Non-Muscle Invasive Bladder Cancer after Protein Corona-Silver Nanoparticles Analysis by SWATH-MS. Nanomaterials, 11(9), 2384. https://doi.org/10.3390/nano11092384