Assessment of New Strategies to Improve the Performance of Antimicrobial Peptides
Abstract
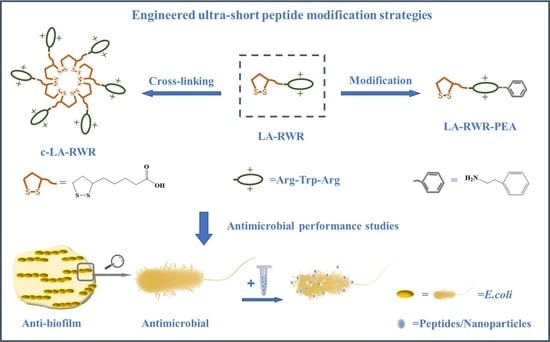
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Peptide Synthesis and Purification
2.3. Crosslinking of AMPs
2.4. Characterization of AMP Nanoparticles
2.5. Minimum Inhibitory Concentration (MIC) Assay
2.6. Stability Evaluation
2.7. Time-Kill Kinetic Assay
2.8. Anti-Biofilm Assay
2.9. Membrane Depolarization Studies
2.10. Membrane Permeability Studies
2.11. Visual Analysis
2.12. Hemolytic Activity Assay
2.13. Cytotoxicity Assay
3. Results
3.1. Characterization of c-LA-RWR/c-LA-RWR-PEA Nanoparticles
3.2. Antimicrobial Activities of Peptides in Vitro
3.3. Biofilm Eradication and Inhibition Activities of Peptides
3.4. Antimicrobial Mechanisms of Peptides
3.4.1. Mechanisms of Membrane Depolarization
3.4.2. Mechanisms of Outer Membrane Permeability
3.5. Visualization of Cell Damage
3.6. In Vivo Safety Evaluation of Peptides
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Appendix A




References
- Yan, Y.; Li, Y.; Zhang, Z.; Wang, X.; Niu, Y.; Zhang, S.; Xu, W.; Ren, C. Advances of peptides for antibacterial applications. Colloids Surf B Biointerfaces 2021, 202, 111682. [Google Scholar] [CrossRef] [PubMed]
- Almaaytah, A.; Qaoud, M.T.; Khalil Mohammed, G.; Abualhaijaa, A.; Knappe, D.; Hoffmann, R.; Al-Balas, Q. Antimicrobial and Antibiofilm Activity of UP-5, an Ultrashort Antimicrobial Peptide Designed Using Only Arginine and Biphenylalanine. Pharmaceuticals 2018, 11, 3. [Google Scholar] [CrossRef] [Green Version]
- Nagarajan, K.; Kumar, V.; Rai, P.K.; Singh, J.; Panda, B.P.; Ghosh, L.K. Assessment of antibacterial activity of smaller chain tripeptides and tetrapeptides. Drug Res. 2014, 64, 623–628. [Google Scholar] [CrossRef] [PubMed]
- Raheem, N.; Straus, S.K. Mechanisms of Action for Antimicrobial Peptides With Antibacterial and Antibiofilm Functions. Front. Microbiol. 2019, 10, 2866. [Google Scholar] [CrossRef] [Green Version]
- Seyfi, R.; Kahaki, F.A.; Ebrahimi, T.; Montazersaheb, S.; Eyvazi, S.; Babaeipour, V.; Tarhriz, V. Antimicrobial Peptides (AMPs): Roles, Functions and Mechanism of Action. Int. J. Pept. Res. Ther. 2019, 26, 1451–1463. [Google Scholar] [CrossRef]
- Molchanova, N.; Hansen, P.R.; Franzyk, H. Advances in Development of Antimicrobial Peptidomimetics as Potential Drugs. Molecules 2017, 22, 1430. [Google Scholar] [CrossRef] [Green Version]
- Yeaman, M.R.; Yount, N.Y. Mechanisms of antimicrobial peptide action and resistance. Pharmacol. Rev. 2003, 55, 27–55. [Google Scholar] [CrossRef] [Green Version]
- Andersson, D.I.; Hughes, D.; Kubicek-Sutherland, J.Z. Mechanisms and consequences of bacterial resistance to antimicrobial peptides. Drug Resist. Updat. 2016, 26, 43–57. [Google Scholar] [CrossRef]
- Sharma, K.; Aaghaz, S.; Shenmar, K.; Jain, R. Short Antimicrobial Peptides. Recent Pat. Antiinfect. Drug Discov. 2018, 13, 12–52. [Google Scholar] [CrossRef]
- Schmidtchen, A.; Pasupuleti, M.; Malmsten, M. Effect of hydrophobic modifications in antimicrobial peptides. Adv. Colloid Interface Sci. 2014, 205, 265–274. [Google Scholar] [CrossRef]
- Jiang, Y.; Chen, Y.; Song, Z.; Tan, Z.; Cheng, J. Recent advances in design of antimicrobial peptides and polypeptides toward clinical translation. Adv. Drug Deliv. Rev. 2021, 170, 261–280. [Google Scholar] [CrossRef] [PubMed]
- Nestor, J.J., Jr. The Medicinal Chemistry of Peptides. Curr. Med. Chem. 2009, 16, 4399–4418. [Google Scholar] [CrossRef] [PubMed]
- Wang, G. Post-Translational Modifications of Natural Antimicrobial Peptides and Strategies for Peptide Engineering. Curr. Biotechnol. 2012, 1, 72–79. [Google Scholar] [CrossRef] [PubMed]
- Gentilucci, L.; Tolomelli, A.; Squassabia, F. Peptides and peptidomimetics in medicine, surgery and biotechnology. Curr. Med. Chem. 2006, 13, 2449–2466. [Google Scholar] [CrossRef]
- Gentilucci, L.; De Marco, R.; Cerisoli, L. Chemical Modifications Designed to Improve Peptide Stability: Incorporation of Non-Natural Amino Acids, Pseudo-Peptide Bonds, and Cyclization. Curr. Pharm. Des. 2010, 16, 3185–3203. [Google Scholar] [CrossRef]
- Diao, L.; Meibohm, B. Pharmacokinetics and Pharmacokinetic–Pharmacodynamic Correlations of Therapeutic Peptides. Clin. Pharmacokinet. 2013, 52, 855–868. [Google Scholar] [CrossRef] [PubMed]
- Das, R.; Gayakvad, B.; Shinde, S.D.; Rani, J.; Jain, A.; Sahu, B. Ultrashort Peptides-A Glimpse into the Structural Modifications and Their Applications as Biomaterials. ACS Appl. Bio. Mater. 2020, 3, 5474–5499. [Google Scholar] [CrossRef]
- Chen, L.; Shen, T.; Liu, Y.; Zhou, J.; Shi, S.; Wang, Y.; Zhao, Z.; Yan, Z.; Liao, C.; Wang, C. Enhancing the antibacterial activity of antimicrobial peptide PMAP-37(F34-R) by cholesterol modification. BMC Vet. Res. 2020, 16, 419. [Google Scholar] [CrossRef] [PubMed]
- Kamysz, E.; Sikorska, E.; Jaskiewicz, M.; Bauer, M.; Neubauer, D.; Bartoszewska, S.; Baranska-Rybak, W.; Kamysz, W. Lipidated Analogs of the LL-37-Derived Peptide Fragment KR12-Structural Analysis, Surface-Active Properties and Antimicrobial Activity. Int. J. Mol. Sci. 2020, 21, 887. [Google Scholar] [CrossRef] [Green Version]
- Richter, F.; Mapfumo, P.; Martin, L.; Solomun, J.I.; Hausig, F.; Frietsch, J.J.; Ernst, T.; Hoeppener, S.; Brendel, J.C.; Traeger, A. Improved gene delivery to K-562 leukemia cells by lipoic acid modified block copolymer micelles. J. Nanobiotechnology 2021, 19, 70. [Google Scholar] [CrossRef] [PubMed]
- Zheng, M.; Zhong, Y.; Meng, F.; Peng, R.; Zhong, Z. Lipoic acid modified low molecular weight polyethylenimine mediates nontoxic and highly potent in vitro gene transfection. Mol. Pharm. 2011, 8, 2434–2443. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Wang, L.; Chen, Y.; Han, H.; Li, Q. Lipoic Acid-Modified Oligoethyleneimine-Mediated miR-34a Delivery to Achieve the Anti-Tumor Efficacy. Molecules 2021, 26, 4827. [Google Scholar] [CrossRef]
- Xie, X.; Zeng, P.; Song, Y.; Zhu, C. Biodegradation and interactions of fosfomycin and α-phenylethylamine in pharmaceutical wastewater. China Environ. Sci. 2014, 34, 2824–2830. [Google Scholar]
- Beckloff, N.; Laube, D.; Castro, T.; Furgang, D.; Park, S.; Perlin, D.; Clements, D.; Tang, H.; Scott, R.W.; Tew, G.N.; et al. Activity of an antimicrobial peptide mimetic against planktonic and biofilm cultures of oral pathogens. Antimicrob. Agents Chemother. 2007, 51, 4125–4132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Z.; Brady, A.; Young, A.; Rasimick, B.; Chen, K.; Zhou, C.; Kallenbach, N.R. Length effects in antimicrobial peptides of the (RW)n series. Antimicrob. Agents Chemother. 2007, 51, 597–603. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.H.; Lu, T.K. Development and Challenges of Antimicrobial Peptides for Therapeutic Applications. Antibiotics 2020, 9, 24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhong, C.; Zhang, F.; Zhu, N.; Zhu, Y.; Yao, J.; Gou, S.; Xie, J.; Ni, J. Ultra-short lipopeptides against gram-positive bacteria while alleviating antimicrobial resistance. Eur. J. Med. Chem. 2021, 212, 113138. [Google Scholar] [CrossRef]
- Roy, R.; Tiwari, M.; Donelli, G.; Tiwari, V. Strategies for combating bacterial biofilms: A focus on anti-biofilm agents and their mechanisms of action. Virulence 2018, 9, 522–554. [Google Scholar] [CrossRef] [Green Version]
- Lin, Q.; Deslouches, B.; Montelaro, R.C.; Di, Y.P. Prevention of ESKAPE pathogen biofilm formation by antimicrobial peptides WLBU2 and LL37. Int. J. Antimicrob. Agents 2018, 52, 667–672. [Google Scholar] [CrossRef]
- Morroni, G.; Simonetti, O.; Brenciani, A.; Brescini, L.; Kamysz, W.; Kamysz, E.; Neubauer, D.; Caffarini, M.; Orciani, M.; Giovanetti, E.; et al. In vitro activity of Protegrin-1, alone and in combination with clinically useful antibiotics, against Acinetobacter baumannii strains isolated from surgical wounds. Med. Microbiol. Immunol. 2019, 208, 877–883. [Google Scholar] [CrossRef]
- Pinto, S.N.; Dias, S.A.; Cruz, A.F.; Mil-Homens, D.; Fernandes, F.; Valle, J.; Andreu, D.; Prieto, M.; Castanho, M.; Coutinho, A.; et al. The mechanism of action of pepR, a viral-derived peptide, against Staphylococcus aureus biofilms. J. Antimicrob. Chemother. 2019, 74, 2617–2625. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Li, Y.; Wang, H.; Zhu, M.; Feng, W.; Liang, G. Enhanced Antibacterial and Anti-Biofilm Activities of Antimicrobial Peptides Modified Silver Nanoparticles. Int. J. Nanomed. 2021, 16, 4831–4846. [Google Scholar] [CrossRef]
- Bessa, L.J.; Manickchand, J.R.; Eaton, P.; Leite, J.; Brand, G.D.; Gameiro, P. Intragenic Antimicrobial Peptide Hs02 Hampers the Proliferation of Single- and Dual-Species Biofilms of P. aeruginosa and S. aureus: A Promising Agent for Mitigation of Biofilm-Associated Infections. Int. J. Mol. Sci. 2019, 20, 3604. [Google Scholar] [CrossRef] [Green Version]
- Gan, B.-H.; Siriwardena, T.N.; Javor, S.; Darbre, T.; Reymond, J.-L. Fluorescence Imaging of Bacterial Killing by Antimicrobial Peptide Dendrimer G3KL. ACS Infect. Dis. 2019, 5, 2164–2173. [Google Scholar] [CrossRef]
- Yasir, M.; Dutta, D.; Willcox, M.D.P. Mode of action of the antimicrobial peptide Mel4 is independent of Staphylococcus aureus cell membrane permeability. PLoS ONE 2019, 14, e0215703. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, F.; Wang, H.; Cao, S.; Jiang, C.; Hou, J. Characterization of antibacterial activity and mechanisms of two linear derivatives of bactenecin. LWT 2019, 107, 89–97. [Google Scholar] [CrossRef]
- Rodrigues de Almeida, N.; Han, Y.; Perez, J.; Kirkpatrick, S.; Wang, Y.; Sheridan, M.C. Design, Synthesis, and Nanostructure-Dependent Antibacterial Activity of Cationic Peptide Amphiphiles. ACS Appl. Mater. Interfaces 2019, 11, 2790–2801. [Google Scholar] [CrossRef] [PubMed]
- Nam, B.H.; Park, E.H.; Shin, E.H.; Kim, Y.O.; Kim, D.G.; Kong, H.J.; Park, J.Y.; Seo, J.K. Development of novel antimicrobial peptides derived from anti-lipopolysaccharide factor of the swimming crab, Portunus trituberculatus. Fish Shellfish Immunol. 2019, 84, 664–672. [Google Scholar] [CrossRef]
- Xiong, W.; Chai, J.; Wu, J.; Tian, M.; Lu, W.; Xu, X. Antibacterial mechanism of Brevinin-2GHk, an antimicrobial peptide from Fejervarya limnocharis skin. J. South. Med. Univ. 2021, 41, 1657–1663. [Google Scholar] [CrossRef]
- Kuzmin, D.V.; Emelianova, A.A.; Kalashnikova, M.B.; Panteleev, P.V.; Balandin, S.V.; Serebrovskaya, E.O.; Belogurova-Ovchinnikova, O.Y.; Ovchinnikova, T.V. Comparative in vitro study on cytotoxicity of recombinant beta-hairpin peptides. Chem. Biol. Drug Des. 2018, 91, 294–303. [Google Scholar] [CrossRef]
- Madanchi, H.; Ebrahimi Kiasari, R.; Seyed Mousavi, S.J.; Johari, B.; Shabani, A.A.; Sardari, S. Design and Synthesis of Lipopolysaccharide-Binding Antimicrobial Peptides Based on Truncated Rabbit and Human CAP18 Peptides and Evaluation of Their Action Mechanism. Probiotics Antimicrob. Proteins 2020, 12, 1582–1593. [Google Scholar] [CrossRef] [PubMed]
- Yasir, M.; Willcox, M.D.P.; Dutta, D. Action of Antimicrobial Peptides against Bacterial Biofilms. Materials 2018, 11, 2468. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wani, N.A.; Singh, G.; Shankar, S.; Sharma, A.; Katoch, M.; Rai, R. Short hybrid peptides incorporating beta- and gamma-amino acids as antimicrobial agents. Peptides 2017, 97, 46–53. [Google Scholar] [CrossRef] [PubMed]
- Zhao, R.; Wang, H.; Ji, T. Biodegradable cationic ε-poly-L-lysine-conjugated polymeric nanoparticles as a new effective antibacterial agent. Sci. Bull. 2015, 60, 216–226. [Google Scholar] [CrossRef]
- Lu, Y.; Wu, Y.; Liang, J.; Libera, M.R.; Sukhishvili, S.A. Self-defensive antibacterial layer-by-layer hydrogel coatings with pH-triggered hydrophobicity. Biomaterials 2015, 45, 64–71. [Google Scholar] [CrossRef]
- Kaneko, T.; Saito, T.; Shobuike, T.; Miyamoto, H.; Matsuda, J.; Fukazawa, K.; Ishihara, K.; Tanaka, S.; Moro, T. 2-Methacryloyloxyethyl Phosphorylcholine Polymer Coating Inhibits Bacterial Adhesion and Biofilm Formation on a Suture: An In Vitro and In Vivo Study. BioMed Res. Int. 2020, 2020, 5639651. [Google Scholar] [CrossRef]
- Velasco-Bolom, J.L.; Corzo, G.; Garduno-Juarez, R. Molecular dynamics simulation of the membrane binding and disruption mechanisms by antimicrobial scorpion venom-derived peptides. J. Biomol. Struct. Dyn. 2018, 36, 2070–2084. [Google Scholar] [CrossRef] [PubMed]













| Peptides | MIC (µg/mL) | |||
|---|---|---|---|---|
| E. coli | S. aureus | C. albicans | MRSA | |
| LA-RWR | 256 | 256 | 128 | 256 |
| c-LA-RWR | 4 | 8 | 8 | 16 |
| LA-RWR-PEA | 2 | 4 | 8 | 8 |
| c-LA-RWR-PEA | 1 | 2 | 4 | 4 |
| Daptomycin | 1 | 1 | 2 | 2 |
| Peptides | pH 6.8 | pH 7.4 (Normal Medium) | pH 8.0 | 100 mM NaCl | 1 mM CaCl2 | 1 mM MgCl2 | 5% Serum |
|---|---|---|---|---|---|---|---|
| LA-RWR | 512 | 256 | 256 | 512 | 512 | 512 | 256 |
| c-LA-RWR | 4 | 4 | 4 | 16 | 8 | 16 | 8 |
| LA-RWR-PEA | 2 | 2 | 2 | 8 | 4 | 8 | 4 |
| c-LA-RWR-PEA | 2 | 1 | 1 | 4 | 4 | 8 | 2 |
| Peptides | Hemolysis Rate (%) Concentration (µg/mL) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 512 | 256 | 128 | 64 | 32 | 16 | 8 | 4 | 2 | |
| LA-RWR | 9.11 | 7.42 | 6.25 | 5.68 | 4.11 | 3.98 | 3.41 | 2.27 | 1.14 |
| c-LA-RWR | 5.45 | 5.03 | 4.68 | 4.27 | 3.11 | 2.7 | 1.68 | 1.14 | 0.57 |
| LA-RWR-PEA | 5.27 | 4.85 | 4.41 | 3.98 | 3.17 | 2.84 | 2.27 | 1.25 | 0.57 |
| c-LA-RWR-PEA | 4.63 | 3.7 | 3.11 | 2.84 | 2.27 | 1.7 | 1 | 0 | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, L.; Liu, H.; Li, X.; Yao, C. Assessment of New Strategies to Improve the Performance of Antimicrobial Peptides. Nanomaterials 2022, 12, 3691. https://doi.org/10.3390/nano12203691
Wang L, Liu H, Li X, Yao C. Assessment of New Strategies to Improve the Performance of Antimicrobial Peptides. Nanomaterials. 2022; 12(20):3691. https://doi.org/10.3390/nano12203691
Chicago/Turabian StyleWang, Lin, Hang Liu, Xinsong Li, and Chen Yao. 2022. "Assessment of New Strategies to Improve the Performance of Antimicrobial Peptides" Nanomaterials 12, no. 20: 3691. https://doi.org/10.3390/nano12203691
APA StyleWang, L., Liu, H., Li, X., & Yao, C. (2022). Assessment of New Strategies to Improve the Performance of Antimicrobial Peptides. Nanomaterials, 12(20), 3691. https://doi.org/10.3390/nano12203691