2.1. Principal Component Analysis
Raman images of human lung epithelial cells exposed to polystyrene submicron (PS) particles were collected with two different step lengths: 100 nm (referred to as the “100 nm image”) and 500 nm (referred to as the “500 nm image”). Principal component analysis (PCA) was used to visualize particles and cells in the images, and also to reduce the number of variables (wavenumbers) prior to applying super-resolution algorithms, by selecting the first 2 principal components.
In PCA, the variance in data is summarized by new, orthogonal variables, so called principal components (PCs) [
8,
10]. The first PC describes the largest variance [
8,
10]. All spectra/pixels in the Raman images are given score values, which are interpreted as concentrations of the corresponding PC [
8,
10]. The loadings are the weights that should be multiplied with the original variables,
i.e., wavenumbers, in order to get the score values [
8,
10]. They should be interpreted together with the scores [
8,
10]. It should however be noted that negative loading values do not imply that the original spectra have peaks with negative values [
8,
10].
Figure 1 shows the loadings of the first PC (PC1) and second PC (PC2) from PCA models of the 100 nm image and the 500 nm image. It is evident that PC1 captures the fluorescence background and an overall intensity variation in the Raman spectra with pronounced contribution from the Raman band at ≈2900 cm
−1 due to ν(C–H) stretching modes, which can be used to visualize the cells in Raman images [
6,
7,
11]. In the 500 nm image, the lowest intensities and lowest score values are observed for the pixels acquired from the cell and the highest intensity and highest score values are observed from pixels from outside of the cell. An opposite response is observed in PC1 for the 100 nm image.
PS is in both PCA models described by PC2, whose loading plots have similarities to a reference spectrum of PS (
Figure 2). In both models, pixels/spectra from PS have low score values in PC2 (apparent as negative bands in
Figure 2b,d). Variables that are correlated may be described by the same PC, irrespective of the origin of their variance. It is therefore possible that PC2 also contains spectral information from other sources than PS. However, the presence of characteristic Raman bands from PS, their relative intensities, which agrees with the relative intensities of the strongest Raman bands from PS (
Figure 2), and the score maps (
Figure 3b,c), which show probable particles, suggest that PC2 represents PS very well. It is worth noting that the spectral quality is very low in single pixel spectra (
Figure 4) and multivariate analysis such as PCA, which considers averaging over many pixels, can therefore be considered as more reliable than an analysis of single Raman bands. Raw spectra showed only small variations in fluorescence background. Spectral pre-treatment, such as baseline correction, was therefore not necessary. The small variation in fluorescence background seems also to be explained by PC1 and not PC2 (see
Figure 2).
It can be seen that the loading plots for the PCA of the 500 nm image are noisier than the corresponding loading plots for the PCA of the 100 nm image. This is because of the comparatively few spectra in the 500 nm image: 3,705 pixels compared to 79,300 pixels in the 100 nm image. It should be mentioned that the measurement time per pixel is the same in both images, which means that the signal to noise ratio per pixel can be considered as equal in both cases. The total measurement time of the 500 nm image is thus much shorter than the total measurement time of the 100 nm image. A question to be answered is if images acquired with long step lengths, i.e., short measurement times, can be sufficiently improved by super-resolution algorithms such that similar information can be extracted from these images as from images acquired with a shorter step length.
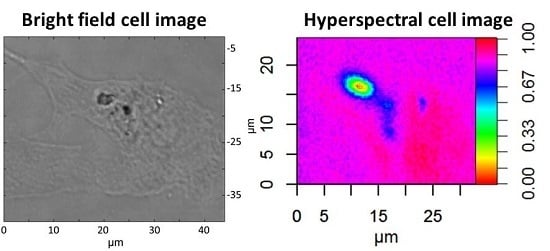
Figure 5 shows the Raman mapped lung epithelial cell. Large particle agglomerates can be observed directly in the optical microscope image, but this image alone is not enough to prove that particles are inside the cell. As a comparison,
Figure 6 shows an optical microscope image of a control cell, which was not exposed to particles. Typically, black dots, which do not represent PS particles, are visible inside cells in optical microscope images of unexposed cells. Thus inspections of optical microscopy images alone cannot be used to distinguish PS particles in cells.
In contrast, the score maps in
Figure 3 show that PS particles are located inside cells.
Figure 3c,d shows the position of PS particles and
Figure 3a,b shows the cell. By overlaying
Figure 3a with
Figure 3c and
Figure 3b with
Figure 3d, it is evident that PS particles are located inside the cell. The position of the largest PS particle agglomerate is at
X ≈ 11.5 µm and
Y ≈ 16.1 µm in the 100 nm image and at
X ≈ 8.5 µm and
Y ≈ 21 µm in the 500 nm image. Some smaller particle agglomerates are located to the right and below the larger agglomerate, and are centered at
X ≈ 16.5 µm (100 nm image) and
X ≈ 11.5 µm (500 nm image). These smaller particle agglomerates and the larger particle agglomerate are not well-separated in the 500 nm image. The particle agglomerate seen at
X ≈ 23 µm,
Y ≈ 13.5 µm in the 100 nm image is also difficult to discern in the 500 nm image, where the background is noisier than in the 100 nm image. Most of the particle agglomerates seen in the score images are much larger than the primary particles, which have a diameter of 300 nm. It is well-known that particles in biological matrices tend to agglomerate and that they are surrounded by proteins [
23]. It is thus expected that particles in a biological system are much larger than corresponding primary particles.
2.2. Super-Resolution and Effect of Regularization Parameter
Score maps were treated with a super-resolution algorithm to remove noise. A general assumption in super-resolution algorithms is that measured, low resolution images (here: score maps) are blurry, noisy, warped and decimated versions of a super-resolution image,
[
17,
18,
19,
20,
21,
22]. For
N low resolution images
, the relationship can be expressed as:
where
denotes decimation,
denotes the geometric warp,
corresponds to the blur function,
is the super-resolution image, and
, is the noise in image
i, respectively [
17,
18,
19,
20,
21,
22]. Since the data was collected in a single mapping experiment, the step length is at high-resolution whereas the optical resolution is low. In our algorithm, the sequence of low-resolution images is constructed by combining subsets of pixels from single mapping acquisitions. The geometric warp and the decimation are thus known. The blur,
i.e., the unresolved details, is here represented by the point spread function (PSF),
i.e., the response of the imaging system to a point source [
17,
18,
19,
20,
21,
22].
can hence be estimated from
, the PSF (
), the geometric warp,
, and the decimation,
, by minimizing the sum of squared residuals [
17,
18,
19,
20,
21,
22]:
The minimum norm solution in Equation (3) is an ill-posed problem, which means that the solution is very sensitive to noise [
17,
18,
19,
20,
21,
22]. To find a robust solution it is therefore necessary to use regularization, which means that a regularization term is added to suppress noise. We have used Tikhonov regularization to calculate the magnitude of
X:
Different forms of regularization terms are possible and Tikhonov prioritize keeping a small gradient in the image, and so here the regularization parameter, α, is used to adjust the smoothing of the super-resolution image,
[
17,
18,
19,
20,
21,
22]. In essence, super-resolution is built into the mathematical formulation posing it to an ill-posed problem, and the Tikhonov choice of regularization term creates a trade-off between sharp contrast and noise removal. It is thus crucial to choose an α-value that gives enough noise removal and at the same time does not alter important information in
.
Detailed descriptions of the super-resolution concept and (Tikhonov) regularization are beyond the scope of this article. We refer the reader to other publications for details of these methods [
17,
18,
19,
20,
21,
22].
The first derivative of intensity profiles,
i.e., line mappings, over the border of a sharp edge between known materials can be used as an approximation of the PSF [
17]. Here we measured across the edge of an Au structure in a 1951 USAF patterned Au/Si reference sample to determine the PSF. The PSF (
Figure 7) was approximated from the measurements of the 1951 USAF patterned Au/Si reference sample by fitting Gaussian functions to the derivatives of the Si Raman band at 520.7 cm
−1 intensity profiles. The experimentally determined PSF map was found to be slightly elliptic, similar to previous reports using same types of Raman microscopes [
17]. The full width at half maximum (FWHM) of the PSF was 1.22 µm (Y-direction) and 1.70 µm (
X-direction). This is much larger than the theoretical spatial resolution according to the Rayleigh criterion (350 nm). It should be remarked that the contrast in the Raman image of the 1951 USAF patterned Au/Si reference sample is much higher than the contrast in the Raman images of particles in cells. The approximated PSF can therefore be expected to be much narrower than the true PSF in the Raman images of cells. The approximated PSF from the Au/Si reference was used as a reasonable approximation since there are no sharp edges in the Raman images of cells from where a reliable PSF can be approximated.
The effect of different α-values was studied in score maps of PC2. Images were treated with the super-resolution algorithm with α varying between 0.01 and 1, and the intensity profiles of the lines at
X = 11.5 µm,
Y = 16.1 µm in the 100 nm image and at
X = 8.5 µm and
Y = 21 µm in the 500 nm image were compared (
Figure 8). All line maps are centered at the largest particle agglomerate. We can expect small α-values to give very unstable solutions to Equation (4) because of its ill-conditioned nature. Images generated with α-values that are too low will thus contain noise and artefacts. This is clearly seen in
Figure 8, where it is shown that α = 0.01 actually gives images with more noise than the original images. α-values, that are too high, on the other hand, produce too much smoothing and thus a loss of important information. The maximum and minimum derivatives of the PC2 line scans are shown in
Table 1. Hard regularization removes high frequency components, such as noise, but also sharp edges. This is illustrated in
Table 1, where it can be seen that the derivatives are impaired with higher α-values. The derivatives of the peak in the intensity profiles are actually worsen in images calculated with high α-values compared to the derivatives in the original score maps. In order to improve the derivatives α = 0.05 is found to be a suitable α-value while maintaining reasonable smoothness in the image. α = 0.01 gives even sharper transition between the particle and the cell in the 100 nm image, but the noise level is evidently increased. To achieve a clear noise removal, it seems, however, that a much higher α-value, α = 0.5, is necessary. Images where thus generated with both α = 0.05 and α = 0.5 to represent enhanced resolution and smoothed images respectively.
The spatial resolution improvement upon application of super-resolution can be assessed by studying the edge of PS intensity lines. Analogous to the FWHM calculation for the PSF determination, edge contrast of PS particles were found by calculating FWHM of the
derivative of the intensity lines at both the positive and negative side.
Table 2 summarizes the calculated FWHM for the original images and the images generated with α = 0.05 and α = 0.5. It is seen that FWHM is not decreased, compared to the original images, by applying super-resolution algorithms except for the resolution in the
X-direction in the 500 nm image, α = 0.05. It can also be noticed that the resolution is similar in the
X direction compared with the line mappings of Au on Si (the 1951 USAF patterned test sample measured to estimate the PSF). In the
Y-direction on the other hand the PS sample has lower edge contrast, which shows the inhomogeneity of the particle agglomerates and possibly resolution “artefacts” of transparent samples [
13].
Table 2 suggests that the resolution is maintained upon application of super-resolution algorithms and the main effect of the super-resolution algorithm to our images is evidently removal of noise.
2.3. Comparison of Super-Resolution Images
The original and super-resolution-treated images of PC2 are shown in
Figure 9. The super-resolution-treated images are slightly smaller than the input images. This is because of the procedure where low-resolution images are formed from the score maps by picking out a subset of pixels. This procedure gives too few pixels with overlapping measurement volumes at the edges of the images to estimate a noise-reduced image by using the super-resolution algorithm.
The smoothing effect of α is evident in
Figure 9. The images generated with α = 0.5 contain much less noise and are smoother than the original images as well as the images generated with α = 0.05. The suspected particles are also easier to discern from the background because of the reduced noise and an increased contrast. The
image quality is however
not improved with α = 0.05. The 500 nm image treated with the super-resolution algorithm α = 0.05 has actually a noisier background than the original 500 nm image. The 100 nm image treated with the super-resolution algorithm and α = 0.05 has a lower contrast than the original image. It is evident that harder regularization than α = 0.05 is necessary to improve the image quality.
Intensity profiles for line mappings at
X ≈ 16.5 (100 nm image) and
X ≈ 11.5 (500 nm image),
i.e., lines centered over the smaller particle agglomerates, were compared to test how well particles positioned next to each other can be separated. The intensity profiles are shown in
Figure 10. The exact number of particles and their diameters are unknown, since it is well-known that particles in biological matrices often are agglomerated and have proteins adsorbed to their surfaces [
23]. A fit of the line mappings to the “true” size of the particles is therefore complicated and a precise answer to the number of closely positioned particles that can be separated before and after application of super-resolution algorithms is hence difficult to achieve.
Figure 10a–c shows broad peaks centered at
ca. 20 µm in the 500 nm images, while
Figure 10d–f shows broad peaks centered at about 10 μm with diffuse shoulders at about 7 μm in the 100 nm images. The intensity profile for the line scan of the original 500 nm image appears to have one broad intensity maxima. Super-resolution with α = 0.05 has only a slightly smoothing effect, and amplifies artefact noise structures (
Figure 10c). Two intensity maxima can possibly also be discerned in the super-resolution 500 nm image with α = 0.5. The penalty in noise reduction is, however, worse spatial resolution. The noise is not reduced for the 100 nm image when applying super-resolution with α = 0.05. The amplitude of the peak is however decreased, and it is thus even more difficult to discern particles in this image. Clear improvements of the 100 nm image are however seen when super-resolution is applied with α = 0.5 (
Figure 10f). The noise is strongly reduced and the two apparent intensity maxima suggest that the line mapping is centered over two large particle agglomerates. Our results show that application of super-resolution algorithm is meaningful only when the sampling step-size is smaller than the object size and that in such cases considerable improvement of noise reduction can be achieved with maintained spatial resolution.
Threshold images (
Figure 11) were constructed from the score maps to study how many particles and agglomerates that can be detected in the images. The information is also summarized in
Table 3, which shows the number and sizes of detected particle agglomerates (≥0.09 µm
2) in the threshold images.
The original 500 nm image is noisy and several single pixels are classified as particles. These particles are not visible in the 100 nm image and they can therefore be regarded as suspected false positives. Super-resolution removes many of these suspected false positives, while other pixels, which are not classified as particles in the original image, become more pronounced with increasing α. In the super-resolution 500 nm image with α = 0.5, four large particle agglomerates are detected, while only two of them are visible in the original 500 nm image. This emphasizes the importance of using a regularization parameter that suppresses noise. It is important to identify possible artefacts in the original images by using e.g., threshold images and line scans before applying super-resolution and to compare the result of regularization with several α-values. Possible artefacts introduced by using insufficient regularization may otherwise give false positives (see
Figure 10c). Taken together this suggests that super-resolution algorithms do not improve the overall quality of our images acquired with long step lengths (500 nm).
In contrast, improvements are clearly seen in the 100 nm image. The smoothing effect of α is also evident. With a small α-value, few, small, particles, or noise, are detected, while a high α-value gives a smoother image, where few, large particles are seen. Only regularization with α = 0.5 enhances the contrast compared to the original image, and the small suspected particle located at X ≈ 23 µm, Y ≈ 13.5 µm is now much easier to discern from the background than in the original 100 nm image. The effect of super-resolution α = 0.05 is very small in the 100 nm image.
In conclusion, our results show that application of super-resolution algorithms does not significantly improve the spatial resolution beyond the theoretical microscopy limit in our biological samples (
Figure 8,
Figure 9,
Figure 10 and
Figure 11). Instead we find that the major advantage of applying super-resolution in these particular applications is noise removal with
maintained spatial resolution, which facilitates reliable detection of particles in complex biological matrices, as visualized by intensity lines over closely positioned particles and by threshold images, where small particles, smaller than can be distinguished from noise in the original images, can be observed. The smoothing effect of the Tikhonov regularization is adjusted by the regularization parameter α: high α-values give smoother images, but may on the other hand give less sharp edges. In this case, the original images are noisy and it is therefore advantageous to use a high α-value in order to detect more particles in the images. Images calculated with super-resolution algorithms using high α-values yield improved contrasts and reduced noise, which facilitates the separation of closely positioned particle agglomerates. The effect is most pronounced in the 100 nm step-size images compared to the 500-nm step-size images acquired with the same measurement time per pixel, which is not surprising since it contains many more pixels than the 500 nm image. The original 500 nm image contains more noise than the 100 nm image and some of the artefacts are enhanced after application of the super-resolution algorithm. To detect as many particles as possible and at the same time avoid noise, it is therefore suggested to collect images with a short step length and thereafter apply super-resolution algorithms to further improve the image quality.
While others have demonstrated image quality enhancements in Raman images by application of super-resolution algorithms in e.g., Si-based materials [
18] and atmospheric inorganic particles [
17], this is the first time a super-resolution algorithm is applied to Raman images of more complex, biological samples. In contrast to Si-based samples, the different components of a Raman image of a cell exposed to particles, e.g., the cell itself or the particles inside the cell, cannot be assessed by integration of a single Raman band, and multivariate techniques are therefore required. In contrast to more stable samples, such as the Si-based samples and particle samples, it is not possible to have long integration times to collect the images of cells, because of the risk of photo damage. We have here demonstrated that the super-resolution algorithm improves the image quality and maintains the image resolution of Raman images in biological samples. Enhanced image quality is of the utmost importance because of the limited measurement time in such samples.