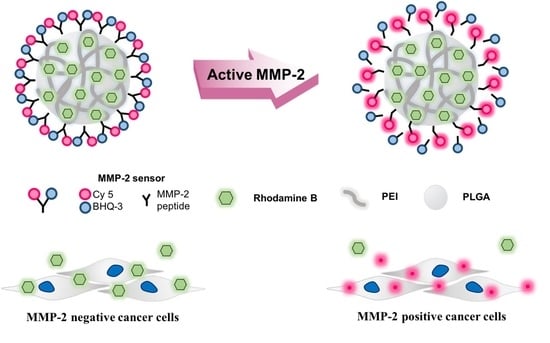
Visualization of MMP-2 Activity Using Dual-Probe Nanoparticles to Detect Potential Metastatic Cancer Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Synthesis and Characterization of MMP-2-Activated Peptide Sensor
2.3. Preparation of PLGA-PEI Nanoparticles
2.4. Conjugation of MMP-2-Activated Peptide Sensor with PLGA-PEI Nanoparticles
2.5. Size, Stability, and Morphology Characterization of MMP-2-PLGA-PEI Nanoparticles
2.6. NIR Fluorescence Signal Recovery of MMP-2-PLGA-PEI Nanoparticles by Active Recombinant Enzyme
2.7. NIR Fluorescence Signal Recovery of MMP-2-PLGA-PEI Nanoparticles by Culture Media and Cell Lysate
2.8. In Vitro Cytotoxicity of MMP-2-PLGA-PEI Nanoparticles
2.9. Confocal Microscopy
2.10. RNA Isolation and RT-PCR
2.11. Statistical Analysis
3. Results and Discussion
3.1. Characterization of the MMP-2-Activated Peptide Sensor
3.2. Characterization of the MMP-2-PLGA-PEI Nanoparticles
3.3. Confirmation of MMP-2 Gene Expression, Cellular Cytotoxicity, and Cell Internalization
3.4. Evaluation of Cellular Active MMP-2 Using MMP-2-PLGA-PEI Nanoparticles
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Kessenbrock, K.; Plaks, V.; Werb, Z. Matrix metalloproteinases: Regulators of the tumor microenvironment. Cell. 2010, 141, 52–67. [Google Scholar] [CrossRef] [PubMed]
- Egeblad, M.; Werb, Z. New functions for the matrix metalloproteinases in cancer progression. Nat. Rev. Cancer 2002, 2, 161–174. [Google Scholar] [CrossRef] [PubMed]
- Huppertz, B.; Kertschanska, S.; Demir, A.Y.; Frank, H.G.; Kaufmann, P. Immunohistochemistry of matrix metalloproteinases (MMP), their substrates, and their inhibitors (TIMP) during trophoblast invasion in the human placenta. Cell. Tissue Res. 1998, 291, 133–148. [Google Scholar] [CrossRef] [PubMed]
- Wysocki, A.B.; Staiano-Coico, L.; Grinnell, F. Wound fluid from chronic leg ulcers contains elevated levels of metalloproteinases MMP-2 and MMP-9. J. Invest. Dermatol. 1993, 101, 64–68. [Google Scholar] [CrossRef] [PubMed]
- Jackson, H.W.; Defamie, V.; Waterhouse, P.; Khokha, R. Timps: Versatile extracellular regulators in cancer. Nat. Rev. Cancer 2017, 17, 38–53. [Google Scholar] [CrossRef] [PubMed]
- Nelson, A.R.; Fingleton, B.; Rothenberg, M.L.; Matrisian, L.M. Matrix metalloproteinases: Biologic activity and clinical implications. J. Clin. Oncol. 2000, 18, 1135–1149. [Google Scholar] [CrossRef] [PubMed]
- Itoh, T.; Tanioka, M.; Yoshida, H.; Yoshioka, T.; Nishimoto, H.; Itohara, S. Reduced angiogenesis and tumor progression in gelatinase a-deficient mice. Cancer Res. 1998, 58, 1048–1051. [Google Scholar] [PubMed]
- Tallant, C.; Marrero, A.; Gomis-Ruth, F.X. Matrix metalloproteinases: Fold and function of their catalytic domains. Biochim. Biophys. Acta 2010, 1803, 20–28. [Google Scholar] [CrossRef] [PubMed]
- Cathcart, J.; Pulkoski-Gross, A.; Cao, J. Targeting matrix metalloproteinases in cancer: Bringing new life to old ideas. Genes Dis. 2015, 2, 26–34. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Wang, Y.; Chen, Z.; Sternlicht, M.D.; Hidalgo, M.; Steffensen, B. Matrix metalloproteinase-2 contributes to cancer cell migration on collagen. Cancer Res. 2005, 65, 130–136. [Google Scholar] [PubMed]
- Dong, W.; Li, H.; Zhang, Y.; Yang, H.; Guo, M.; Li, L.; Liu, T. Matrix metalloproteinase 2 promotes cell growth and invasion in colorectal cancer. Acta Biochim. Biophys. Sin. (Shanghai) 2011, 43, 840–848. [Google Scholar] [CrossRef] [PubMed]
- Munoz-Najar, U.M.; Neurath, K.M.; Vumbaca, F.; Claffey, K.P. Hypoxia stimulates breast carcinoma cell invasion through MT1-MMP and MMP-2 activation. Oncogene 2006, 25, 2379–2392. [Google Scholar] [CrossRef] [PubMed]
- Roomi, M.W.; Monterrey, J.C.; Kalinovsky, T.; Rath, M.; Niedzwiecki, A. Patterns of MMP-2 and MMP-9 expression in human cancer cell lines. Oncol. Rep. 2009, 21, 1323–1333. [Google Scholar] [PubMed]
- Zheng, H.; Takahashi, H.; Murai, Y.; Cui, Z.; Nomoto, K.; Niwa, H.; Tsuneyama, K.; Takano, Y. Expressions of MMP-2, MMP-9 and VEGF are closely linked to growth, invasion, metastasis and angiogenesis of gastric carcinoma. Anticancer Res. 2006, 26, 3579–3583. [Google Scholar] [PubMed]
- Hu, X.; Beeton, C. Detection of functional matrix metalloproteinases by zymography. J. Vis. Exp. 2010. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.; Choi, S.J.; Park, K.; Park, J.W.; Kim, K.; Choi, K.; Yoon, S.Y.; Youn, I. Detection of active matrix metalloproteinase-3 in serum and fibroblast-like synoviocytes of collagen-induced arthritis mice. Bioconjug. Chem. 2013, 24, 1068–1074. [Google Scholar] [CrossRef] [PubMed]
- Leber, T.M.; Balkwill, F.R. Zymography: A single-step staining method for quantitation of proteolytic activity on substrate gels. Anal. Biochem. 1997, 249, 24–28. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Ryu, J.H.; Park, K.; Lee, A.; Lee, S.Y.; Youn, I.C.; Ahn, C.H.; Yoon, S.M.; Myung, S.J.; Moon, D.H. , et al. Polymeric nanoparticle-based activatable near-infrared nanosensor for protease determination in vivo. Nano Lett. 2009, 9, 4412–4416. [Google Scholar] [CrossRef] [PubMed]
- Aryal, S.; Key, J.; Stigliano, C.; Ananta, J.S.; Zhong, M.; Decuzzi, P. Engineered magnetic hybrid nanoparticles with enhanced relaxivity for tumor imaging. Biomaterials 2013, 34, 7725–7732. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.Y.; Huang, L.; Lee, K.M.; Li, K.; Kumta, S.M. Alendronate regulates cell invasion and MMP-2 secretion in human osteosarcoma cell lines. Pediatr. Blood Cancer 2004, 42, 410–415. [Google Scholar] [CrossRef] [PubMed]
- Davidson, B.; Goldberg, I.; Kopolovic, J.; Lerner-Geva, L.; Gotlieb, W.H.; Ben-Baruch, G.; Reich, R. MMP-2 and TIMP-2 expression correlates with poor prognosis in cervical carcinoma—A clinicopathologic study using immunohistochemistry and mRNA in situ hybridization. Gynecol. Oncol. 1999, 73, 372–382. [Google Scholar] [CrossRef] [PubMed]
- Talvensaari-Mattila, A.; Paakko, P.; Turpeenniemi-Hujanen, T. MMP-2 positivity and age less than 40 years increases the risk for recurrence in premenopausal patients with node-positive breast carcinoma. Breast Cancer Res. Treat. 1999, 58, 287–293. [Google Scholar] [CrossRef] [PubMed]
- Rundhaug, J.E. Matrix metalloproteinases and angiogenesis. J. Cell. Mol. Med. 2005, 9, 267–285. [Google Scholar] [CrossRef] [PubMed]
- Fridman, R.; Toth, M.; Pena, D.; Mobashery, S. Activation of progelatinase B (MMP-9) by gelatinase A (MMP-2). Cancer Res. 1995, 55, 2548–2555. [Google Scholar] [PubMed]
- Beck, I.M.; Muller, M.; Mentlein, R.; Sadowski, T.; Mueller, M.S.; Paus, R.; Sedlacek, R. Matrix metalloproteinase-19 expression in keratinocytes is repressed by transcription factors Tst-1 and Skn-1a: Implications for keratinocyte differentiation. J. Invest. Dermatol. 2007, 127, 1107–1114. [Google Scholar] [CrossRef] [PubMed]
- Nawrocki Raby, B.; Polette, M.; Gilles, C.; Clavel, C.; Strumane, K.; Matos, M.; Zahm, J.M.; Van Roy, F.; Bonnet, N.; Birembaut, P. Quantitative cell dispersion analysis: New test to measure tumor cell aggressiveness. Int. J. Cancer 2001, 93, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Bivas-Benita, M.; Romeijn, S.; Junginger, H.E.; Borchard, G. PLGA-PEI nanoparticles for gene delivery to pulmonary epithelium. Eur J. Pharm Biopharm 2004, 58, 1–6. [Google Scholar] [CrossRef] [PubMed]





© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, A.; Kim, S.H.; Lee, H.; Kim, B.; Kim, Y.S.; Key, J. Visualization of MMP-2 Activity Using Dual-Probe Nanoparticles to Detect Potential Metastatic Cancer Cells. Nanomaterials 2018, 8, 119. https://doi.org/10.3390/nano8020119
Lee A, Kim SH, Lee H, Kim B, Kim YS, Key J. Visualization of MMP-2 Activity Using Dual-Probe Nanoparticles to Detect Potential Metastatic Cancer Cells. Nanomaterials. 2018; 8(2):119. https://doi.org/10.3390/nano8020119
Chicago/Turabian StyleLee, Aeju, Sung Hoon Kim, Hyun Lee, Bohee Kim, Yoon Suk Kim, and Jaehong Key. 2018. "Visualization of MMP-2 Activity Using Dual-Probe Nanoparticles to Detect Potential Metastatic Cancer Cells" Nanomaterials 8, no. 2: 119. https://doi.org/10.3390/nano8020119
APA StyleLee, A., Kim, S. H., Lee, H., Kim, B., Kim, Y. S., & Key, J. (2018). Visualization of MMP-2 Activity Using Dual-Probe Nanoparticles to Detect Potential Metastatic Cancer Cells. Nanomaterials, 8(2), 119. https://doi.org/10.3390/nano8020119