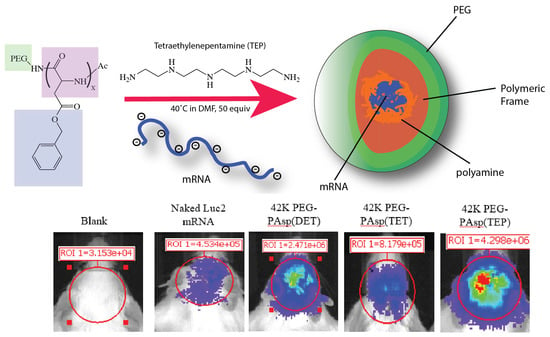
Preparation of Messenger RNA Nanomicelles via Non-Cytotoxic PEG-Polyamine Nanocomplex for Intracerebroventicular Delivery: A Proof-of-Concept Study in Mouse Models
Abstract
:1. Introduction
2. Methods and Materials
2.1. Synthesis of Block Copolymer and NMR Analysis
2.2. Construction of Vector for In Vitro Transcription (IVT) and Preparation of IVT mRNA
2.3. Intracerebrovetricular (ICV) Administration
2.4. Luciferase Expression Measurement by Bioluminescence Assay
2.5. Histological Examination
2.6. Quantitative REAL Time PCR (qRT-PCR) to Examine the Immune Response after Nanomicelle ICV Administration
2.7. Statistical Analysis
3. Results and Discussion
3.1. Synthesis of Block Copolymer and Preparation of Polyplex Nanomicelles
3.2. NMR Analysis of Block Copolymer
3.3. Physical Property Analysis of Polyplex Nanomicelles
3.4. The Proof-of-Concept (POC) Study to Demonstrate the Polyplex Nanomicelles Mediated mRNA Expression in Mouse Brains
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Luo, D.; Saltzman, W.M. Synthetic DNA delivery systems. Nat. Biotechnol. 2000, 18, 33–37. [Google Scholar] [CrossRef] [PubMed]
- Pack, D.W.; Hoffman, A.S.; Pun, S.; Stayton, P.S. Design and development of polymers for gene delivery. Nat. Rev. Drug Discov. 2005, 4, 581–593. [Google Scholar] [CrossRef] [PubMed]
- Knudsen, K.B.; Northeved, H.; Kumar, P.E.K.; Permin, A.; Gjetting, T.; Andresen, T.L.; Larsen, S.; Wegener, K.M.; Lykkesfeldt, J.; Jantzen, K.; et al. In vivo toxicity of cationic micelles and liposomes. Nanomed. Nanotechnol. Biol. Med. 2015, 11, 467–477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roursgaard, M.; Knudsen, K.B.; Northeved, H.; Persson, M.; Christensen, T.; Kumar, P.E.K.; Permin, A.; Andresen, T.L.; Gjetting, T.; Lykkesfeldt, J.; et al. In vitro toxicity of cationic micelles and liposomes in cultured human hepatocyte (hepg2) and lung epithelial (a549) cell lines. Toxicol. Vitro 2016, 36, 164–171. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.J.; Li, H.; Ma, M.S.; Fu, J.; Dong, Y.Z.; Guo, P.X. Size, shape, and sequence-dependent immunogenicity of rna nanoparticles. Mol. Ther.-Nucleic Acids 2017, 9, 399–408. [Google Scholar] [CrossRef] [PubMed]
- Angelov, B.; Garamus, V.M.; Drechsler, M.; Angelova, A. Structural analysis of nanoparticulate carriers for encapsulation of macromolecular drugs. J. Mol. Liq. 2017, 235, 83–89. [Google Scholar] [CrossRef]
- Angelova, A.; Garamus, V.M.; Angelov, B.; Tian, Z.F.; Li, Y.W.; Zou, A.H. Advances in structural design of lipid-based nanoparticle carriers for delivery of macromolecular drugs, phytochemicals and anti-tumor agents. Adv. Colloid Interface Sci. 2017, 249, 331–345. [Google Scholar] [CrossRef]
- Angelov, B.; Angelova, A.; Filippov, S.K.; Narayanan, T.; Drechsler, M.; Stepanek, P.; Couvreur, P.; Lesieur, S. DNA/fusogenic lipid nanocarrier assembly: Millisecond structural dynamics. J. Phys. Chem. Lett. 2013, 4, 1959–1964. [Google Scholar] [CrossRef]
- Uchida, H.; Miyata, K.; Oba, M.; Ishii, T.; Suma, T.; Itaka, K.; Nishiyama, N.; Kataoka, K. Odd-even effect of repeating aminoethylene units in the side chain of n-substituted polyaspartamides on gene transfection profiles. J. Am. Chem. Soc. 2011, 133, 15524–15532. [Google Scholar] [CrossRef]
- Uchida, H.; Itaka, K.; Nomoto, T.; Ishii, T.; Suma, T.; Ikegami, M.; Miyata, K.; Oba, M.; Nishiyama, N.; Kataoka, K. Modulated protonation of side chain aminoethylene repeats in n-substituted polyaspartamides promotes mrna transfection. J. Am. Chem. Soc. 2014, 136, 12396–12405. [Google Scholar] [CrossRef]
- Aini, H.; Itaka, K.; Fujisawa, A.; Uchida, H.; Uchida, S.; Fukushima, S.; Kataoka, K.; Saito, T.; Chung, U.; Ohba, S. Messenger rna delivery of a cartilage-anabolic transcription factor as a disease-modifying strategy for osteoarthritis treatment. Sci. Rep. 2016, 6, 18743. [Google Scholar] [CrossRef] [PubMed]
- Nagata, K.; Itaka, K.; Baba, M.; Uchida, S.; Ishii, T.; Kataoka, K. Muscle-targeted hydrodynamic gene introduction of insulin-like growth factor-1 using polyplex nanomicelle to treat peripheral nerve injury. J. Control. Release 2014, 183, 27–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Varkouhi, A.K.; Scholte, M.; Storm, G.; Haisma, H.J. Endosomal escape pathways for delivery of biologicals. J. Control. Release 2011, 151, 220–228. [Google Scholar] [CrossRef] [PubMed]
- Gu, J.X.; Cheng, W.P.; Liu, J.G.; Lo, S.Y.; Smith, D.; Qu, X.Z.; Yang, Z.Z. Ph-triggered reversible "stealth" polycationic micelles. Biomacromolecules 2008, 9, 255–262. [Google Scholar] [CrossRef] [PubMed]
- Stasko, N.A.; Johnson, C.B.; Schoenfisch, M.H.; Johnson, T.A.; Holmuhamedov, E.L. Cytotoxicity of polypropylenimine dendrimer conjugates on cultured endothelial cells. Biomacromolecules 2007, 8, 3853–3859. [Google Scholar] [CrossRef] [PubMed]
- Amoozgar, Z.; Yeo, Y. Recent advances in stealth coating of nanoparticle drug delivery systems. Wiley Interdiscip. Rev.-Nanomed. Nanobiotechnol. 2012, 4, 219–233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gref, R.; Luck, M.; Quellec, P.; Marchand, M.; Dellacherie, E.; Harnisch, S.; Blunk, T.; Muller, R.H. ‘Stealth’ corona-core nanoparticles surface modified by polyethylene glycol (peg): Influences of the corona (peg chain length and surface density) and of the core composition on phagocytic uptake and plasma protein adsorption. Colloids Surf. B-Biointerfaces 2000, 18, 301–313. [Google Scholar] [CrossRef]
- Jokerst, J.V.; Lobovkina, T.; Zare, R.N.; Gambhir, S.S. Nanoparticle pegylation for imaging and therapy. Nanomedicine 2011, 6, 715–728. [Google Scholar] [CrossRef]
- Lin, C.Y.; Perche, F.; Ikegami, M.; Uchida, S.; Kataoka, K.; Itaka, K. Messenger rna-based therapeutics for brain diseases: An animal study for augmenting clearance of beta-amyloid by intracerebral administration of neprilysin mrna loaded in polyplex nanomicelles. J. Control. Release 2016, 235, 268–275. [Google Scholar] [CrossRef]
- Uchida, S.; Itaka, K.; Uchida, H.; Hayakawa, K.; Ogata, T.; Ishii, T.; Fukushima, S.; Osada, K.; Kataoka, K. In vivo messenger rna introduction into the central nervous system using polyplex nanomicelle. PLoS ONE 2013, 8, e56220. [Google Scholar] [CrossRef] [PubMed]
- Holtkamp, S.; Kreiter, S.; Selmi, A.; Simon, P.; Koslowski, M.; Huber, C.; Tureci, O.; Sahin, U. Modification of antigen-encoding rna increases stability, translational efficacy, and t-cell stimulatory capacity of dendritic cells. Blood 2006, 108, 4009–4017. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Zabner, J.; Deering, C.; Launspach, J.; Shao, J.; Bodner, M.; Jolly, D.J.; Davidson, B.L.; McCray, P.B., Jr. Increasing epithelial junction permeability enhances gene transfer to airway epithelia in vivo. Am. J. Respir. Cell Mol. Biol. 2000, 22, 129–138. [Google Scholar] [CrossRef] [PubMed]
- Itaka, K.; Ishii, T.; Hasegawa, Y.; Kataoka, K. Biodegradable polyamino acid-based polycations as safe and effective gene carrier minimizing cumulative toxicity. Biomaterials 2010, 31, 3707–3714. [Google Scholar] [CrossRef] [PubMed]
- Perlmutter, R.M.; Marth, J.D.; Ziegler, S.F.; Garvin, A.M.; Pawar, S.; Cooke, M.P.; Abraham, K.M. Specialized protein tyrosine kinase proto-oncogenes in hematopoietic cells. Biochim. Biophys. Acta 1989, 948, 245–262. [Google Scholar] [CrossRef]
- Perera, T.; Marzilli, P.A.; Fronczek, F.R.; Marzilli, L.G. Nh nmr shifts of new structurally characterized fac- re(co)(3)(polyamine) (n+) complexes probed via outer-sphere hydrogen-bonding interactions to anions, including the paramagnetic (rebr6)-br-iv (2-) anion. Inorg. Chem. 2010, 49, 5560–5572. [Google Scholar] [CrossRef] [PubMed]
- Kanayama, N.; Fukushima, S.; Nishiyama, N.; Itaka, K.; Jang, W.D.; Miyata, K.; Yamasaki, Y.; Chung, U.I.; Kataoka, K. A peg-based biocompatible block catiomer with high buffering capacity for the construction of polyplex micelles showing efficient gene transfer toward primary cells. ChemMedChem 2006, 1, 439–444. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Ishii, T.; Zheng, M.; Watanabe, S.; Toh, K.; Matsumoto, Y.; Nishiyama, N.; Miyata, K.; Kataoka, K. Multifunctional polyion complex micelle featuring enhanced stability, targetability, and endosome escapability for systemic sirna delivery to subcutaneous model of lung cancer. Drug Deliv. Transl. Res. 2014, 4, 50–60. [Google Scholar] [CrossRef]
- Bolton, P.H.; Kearns, D.R. Hydrogen-bonding interactions of polyamines with 2′ oh of rna. Nucleic Acids Res. 1978, 5, 1315–1324. [Google Scholar] [CrossRef]
- Meneksedag-Erol, D.; Kizhakkedathu, J.N.; Tang, T.; Uludag, H. Molecular dynamics simulations on nucleic acid binding polymers designed to arrest thrombosis. ACS Appl. Mater. Interfaces 2018, 10, 28399–28411. [Google Scholar] [CrossRef]






© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chan, L.Y.; Khung, Y.L.; Lin, C.-Y. Preparation of Messenger RNA Nanomicelles via Non-Cytotoxic PEG-Polyamine Nanocomplex for Intracerebroventicular Delivery: A Proof-of-Concept Study in Mouse Models. Nanomaterials 2019, 9, 67. https://doi.org/10.3390/nano9010067
Chan LY, Khung YL, Lin C-Y. Preparation of Messenger RNA Nanomicelles via Non-Cytotoxic PEG-Polyamine Nanocomplex for Intracerebroventicular Delivery: A Proof-of-Concept Study in Mouse Models. Nanomaterials. 2019; 9(1):67. https://doi.org/10.3390/nano9010067
Chicago/Turabian StyleChan, Long Yi, Yit Lung Khung, and Chin-Yu Lin. 2019. "Preparation of Messenger RNA Nanomicelles via Non-Cytotoxic PEG-Polyamine Nanocomplex for Intracerebroventicular Delivery: A Proof-of-Concept Study in Mouse Models" Nanomaterials 9, no. 1: 67. https://doi.org/10.3390/nano9010067
APA StyleChan, L. Y., Khung, Y. L., & Lin, C. -Y. (2019). Preparation of Messenger RNA Nanomicelles via Non-Cytotoxic PEG-Polyamine Nanocomplex for Intracerebroventicular Delivery: A Proof-of-Concept Study in Mouse Models. Nanomaterials, 9(1), 67. https://doi.org/10.3390/nano9010067