Knockdown of microRNA-135b in Mammary Carcinoma by Targeted Nanodiamonds: Potentials and Pitfalls of In Vivo Applications
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
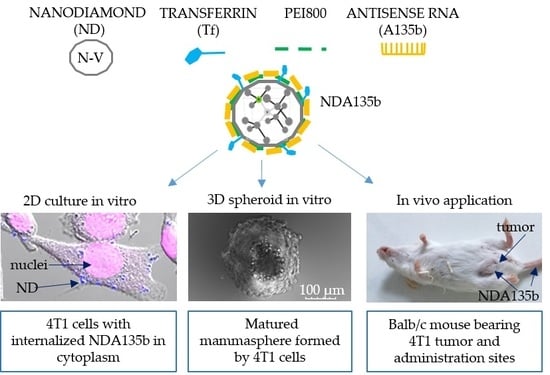
2.1.1. Nanodiamond Complex (NDA135b) Preparation
2.1.2. NDA135b Complex Characterization
2.1.3. Cell Cultures and Transfection
2.1.4. Ex-Vivo Primary Cells Culture and Their Tracking
2.1.5. Quantitative Real-Time-PCR Analysis
2.1.6. Lactate dehydrogenase (LDH) Assay
2.1.7. Flow Cytometry
2.1.8. Confocal Imaging
2.1.9. Animal Model and In Vivo NDA135b Application
2.1.10. Spectrometer Measurements of NDA135b in Fluids
2.1.11. Whole Tissue Imaging
2.1.12. Statistical Analysis
3. Results
3.1. Characterization of the NDA135b Complex
3.2. Effective Delivery of NDA135b into Adherent Breast Cancer Cells
3.3. Specific Internalization of NDA135b into Breast Cancer Cells Spheroids Co-Cultured with Peritoneal Lavage Cells
3.4. Local and Systemic In Vivo Application of Targeted NDA135b
3.5. Accumulation of Non-Internalized NDA135b Complex in Key Tissues after In Vivo Applications
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A

Appendix B


Appendix C

References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bandrés, E.; Cubedo, E.; Agirre, X.; Malumbres, R.; Zárate, R.; Ramirez, N.; Abajo, A.; Navarro, A.; Moreno, I.; Monzó, M.; et al. Identification by Real-time PCR of 13 mature microRNAs differentially expressed in colorectal cancer and non-tumoral tissues. Mol. Cancer 2006, 5, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Tong, A.W.; Fulgham, P.; Jay, C.; Chen, P.; Khalil, I.; Liu, S.; Senzer, N.; Eklund, A.C.; Han, J.; Nemunaitis, J. MicroRNA profile analysis of human prostate cancers. Cancer Gene Ther. 2009, 16, 206–216. [Google Scholar] [CrossRef] [PubMed]
- Lowery, A.J.; Miller, N.; Devaney, A.; McNeill, R.E.; Davoren, P.A.; Lemetre, C.; Benes, V.; Schmidt, S.; Blake, J.; Ball, G.; et al. MicroRNA signatures predict oestrogen receptor, progesterone receptor and HER2/neu receptor status in breast cancer. Breast Cancer Res. 2009, 11, R27. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.L.; Chang, D.C.; Ying, S.Y. Isolation and identification of gene-specific microRNAs. Methods Mol. Biol. 2013, 936, 271–278. [Google Scholar] [PubMed]
- Hayward, S.L.; Francis, D.M.; Kholmatov, P.; Kidambi, S. Targeted Delivery of MicroRNA125a-5p by Engineered Lipid Nanoparticles for the Treatment of HER2 Positive Metastatic Breast Cancer. J. Biomed. Nanotechnol. 2016, 12, 554–568. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, N.; Arora, S.; Deshmukh, S.K.; Singh, S.; Marimuthu, S.; Singh, A.P. Exploiting Nanotechnology for the Development of MicroRNA-Based Cancer Therapeutics. J. Biomed. Nanotechnol. 2016, 12, 28–42. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zhang, J.; Wang, Y.; Chen, M. Hyaluronic acid-coated PEI-PLGA nanoparticles mediated co-delivery of doxorubicin and miR-542-3p for triple negative breast cancer therapy. Nanomedicine 2016, 12, 411–420. [Google Scholar] [CrossRef] [PubMed]
- Zhong, S.; Chen, X.; Wang, D.; Zhang, X.; Shen, H.; Yang, S.; Lv, M.; Tang, J.; Zhao, J. MicroRNA expression profiles of drug-resistance breast cancer cells and their exosomes. Oncotarget 2016, 7, 19601–19609. [Google Scholar] [CrossRef]
- Faklaris, O.; Joshi, V.; Irinopoulou, T.; Tauc, P.; Sennour, M.; Girard, H.; Gesset, C.; Arnault, J.C.; Thorel, A.; Boudou, J.P.; et al. Photoluminescent diamond nanoparticles for cell labeling: Study of the uptake mechanism in mammalian cells. ACS Nano 2009, 3, 3955–3962. [Google Scholar] [CrossRef]
- Alhaddad, A.; Durieu, C.; Dantelle, G.; Le Cam, E.; Malvy, C.; Treussart, F.; Bertrand, J.R. Influence of the internalization pathway on the efficacy of siRNA delivery by cationic fluorescent nanodiamonds in the Ewing sarcoma cell model. PLoS ONE 2012, 7, e52207. [Google Scholar] [CrossRef] [PubMed]
- Petrakova, V.; Benson, V.; Buncek, M.; Fiserova, A.; Ledvina, M.; Stursa, J.; Cigler, P.; Nesladek, M. Imaging of transfection and intracellular release of intact, non-labeled DNA using fluorescent nanodiamonds. Nanoscale 2016, 8, 12002–12012. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lukowski, S.; Neuhoferova, E.; Kinderman, M.; Krivohlava, R.; Mineva, A.; Petrakova, V.; Benson, V. Fluorescent Nanodiamonds are Efficient, Easy-to-Use Cyto-Compatible Vehicles for Monitored Delivery of Non-Coding Regulatory RNAs. J. Biomed. Nanotechnol. 2018, 14, 946–958. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.R.; Lee, H.Y.; Chen, K.; Chang, C.C.; Tsai, D.S.; Fu, C.C.; Lim, T.S.; Tzeng, Y.K.; Fang, C.Y.; Han, C.C.; et al. Mass production and dynamic imaging of fluorescent nanodiamonds. Nat. Nanotechnol. 2008, 3, 284–288. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.Q.; Chen, M.; Lam, R.; Xu, X.; Osawa, E.; Ho, D. Polymer-functionalized nanodiamond platforms as vehicles for gene delivery. ACS Nano 2009, 3, 2609–2616. [Google Scholar] [CrossRef] [PubMed]
- Ho, D.; Wang, C.H.; Chow, E.K. Nanodiamonds: The intersection of nanotechnology, drug development, and personalized medicine. Sci. Adv. 2015, 1, e1500439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, T.; Perona Martínez, F.; Storm, I.M.; Rombouts, W.; Sprakel, J.; Schirhagl, R.; de Vries, R. Recombinant Protein Polymers for Colloidal Stabilization and Improvement of Cellular Uptake of Diamond Nanosensors. Anal. Chem. 2017, 89, 12812–12820. [Google Scholar] [CrossRef] [PubMed]
- Chipaux, M.; van der Laan, K.J.; Hemelaar, S.R.; Hasani, M.; Zheng, T.; Schirhagl, R. Nanodiamonds and Their Applications in Cells. Small 2018, 14, e1704263. [Google Scholar] [CrossRef] [PubMed]
- Hsiao, W.W.; Hui, Y.Y.; Tsai, P.C.; Chang, H.C. Fluorescent Nanodiamond: A Versatile Tool for Long-Term Cell Tracking, Super-Resolution Imaging, and Nanoscale Temperature Sensing. Acc. Chem. Res. 2016, 49, 400–407. [Google Scholar] [CrossRef] [PubMed]
- Mohan, N.; Chen, C.S.; Hsieh, H.H.; Wu, Y.C.; Chang, H.C. In vivo imaging and toxicity assessments of fluorescent nanodiamonds in Caenorhabditis elegans. Nano Lett. 2010, 10, 3692–3699. [Google Scholar] [CrossRef]
- Van der Laan, K.; Hasani, M.; Zheng, T.; Schirhagl, R. Nanodiamonds for In Vivo Applications. Small 2018, 14, e1703838. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Chen, Y.; Liu, J.H.; Wang, H.; Liu, Y. Biodistribution and fate of nanodiamonds in vivo. Diam. Relat. Mater. 2009, 18, 95–100. [Google Scholar] [CrossRef]
- Vaijayanthimala, V.; Cheng, P.Y.; Yeh, S.H.; Liu, K.K.; Hsiao, C.H.; Chao, J.I.; Chang, H.C. The long-term stability and biocompatibility of fluorescent nanodiamond as an in vivo contrast agent. Biomaterials 2012, 33, 7794–7802. [Google Scholar] [CrossRef] [PubMed]
- Tsai, L.W.; Lin, Y.C.; Perevedentseva, E.; Lugovtsov, A.; Priezzhev, A.; Cheng, C.L. Nanodiamonds for Medical Applications: Interaction with Blood in Vitro and in Vivo. Int. J. Mol. Sci. 2016, 17, 1111. [Google Scholar] [CrossRef] [PubMed]
- Hua, K.; Jin, J.; Zhao, J.; Song, J.; Song, H.; Li, D.; Maskey, N.; Zhao, B.; Wu, C.; Xu, H.; et al. miR-135b, upregulated in breast cancer, promotes cell growth and disrupts the cell cycle by regulating LATS2. Int. J. Oncol. 2016, 48, 1791–2423. [Google Scholar] [CrossRef] [PubMed]
- Petrakova, V.; Rehor, I.; Stursa, J.; Ledvina, M.; Nesladek, M.; Cigler, P. Charge-sensitive fluorescent nanosensors created from nanodiamonds. Nanoscale 2015, 7, 12307–12311. [Google Scholar] [CrossRef]
- Upreti, M.; Jamshidi-Parsian, A.; Koonce, N.A.; Webber, J.S.; Sharma, S.K.; Asea, A.A.; Mader, M.J.; Griffin, R.J. Tumor-Endothelial Cell Three-dimensional Spheroids: New Aspects to Enhance Radiation and Drug Therapeutics. Transl. Oncol. 2011, 4, 365–376. [Google Scholar] [CrossRef]
- Longmire, M.; Choyke, P.L.; Kobayashi, H. Clearance properties of nano-sized particles and molecules as imaging agents: Considerations and caveats. Nanomed. Lond. 2008, 3, 703–717. [Google Scholar] [CrossRef]
- Fang, J.; Nakamura, H.; Maeda, H. The EPR effect: Unique features of tumor blood vessels for drug delivery, factors involved, and limitations and augmentation of the effect. Adv. Drug Deliv. Rev. 2011, 63, 136–151. [Google Scholar] [CrossRef]
- Yu, M.; Zheng, J. Clearance Pathways and Tumor Targeting of Imaging Nanoparticles. ACS Nano 2015, 9, 6655–6674. [Google Scholar] [CrossRef] [Green Version]
- Turcheniuk, K.; Mochalin, V.N. Biomedical applications of nanodiamond (Review). Nanotechnology 2017, 28, 252001. [Google Scholar] [CrossRef]
- Yu, S.J.; Kang, M.W.; Chang, H.C.; Chen, K.M.; Yu, Y.C. Bright fluorescent nanodiamonds: No photobleaching and low cytotoxicity. J. Am. Chem. Soc. 2005, 127, 17604–17605. [Google Scholar] [CrossRef]
- Liu, K.K.; Cheng, C.L.; Chang, C.C.; Chao, J.I. Biocompatible and detectable carboxylated nanodiamond on human cell. Nanotechnology 2007, 18, 325102. [Google Scholar] [CrossRef]
- Blaber, S.P.; Hill, C.J.; Webster, R.A.; Say, J.M.; Brown, L.J.; Wang, S.C.; Vesey, G.; Herbert, B.R. Effect of labeling with iron oxide particles or nanodiamonds on the functionality of adipose-derived mesenchymal stem cells. PLoS ONE 2013, 8, e52997. [Google Scholar] [CrossRef]
- Hsu, T.C.; Liu, K.K.; Chang, H.C.; Hwang, E.; Chao, J.I. Labeling of neuronal differentiation and neuron cells with biocompatible fluorescent nanodiamonds. Sci. Rep. 2014, 4, 5004–5015. [Google Scholar] [CrossRef]
- Feng, Q.; Liu, Y.; Huang, J.; Chen, K.; Xiao, K. Uptake, distribution, clearance, and toxicity of iron oxide nanoparticles with different sizes and coatings. Sci. Rep. 2018, 8, 2082–2095. [Google Scholar] [CrossRef]
- Daniels, T.R.; Bernabeu, E.; Rodríguez, J.A.; Patel, S.; Kozman, M.; Chiappetta, D.A.; Holler, E.; Ljubimova, J.Y.; Helguera, G.; Penichet, M.L. The transferrin receptor and the targeted delivery of therapeutic agents against cancer. Biochim. Biophys. Acta 2012, 1820, 291–317. [Google Scholar] [CrossRef] [Green Version]
- Hemelaar, S.R.; Nagl, A.; Bigot, F.; Rodríguez-García, M.M.; de Vries, M.P.; Chipaux, M.; Schirhagl, R. The interaction of fluorescent nanodiamond probes with cellular media. Mikrochim. Acta 2017, 184, 1001–1009. [Google Scholar] [CrossRef] [Green Version]
- Yuan, Y.; Wang, X.; Jia, G. Pulmonary toxicity and translocation of nanodiamond in mice. Diam. Relat. Mater. 2010, 19, 291–300. [Google Scholar] [CrossRef]
- Muñoz, L.E.; Bilyy, R.; Biermann, M.H.; Kienhöfer, D.; Maueröder, C.; Hahn, J.; Brauner, J.M.; Weidner, D.; Chen, J.; Scharin-Mehlmann, M.; et al. Nanoparticles size-dependently initiate self-limiting NETosis-driven inflammation. Proc. Natl. Acad. Sci. USA 2016, 113, 5856–5865. [Google Scholar] [CrossRef]
- Pham, N.B.; Ho, T.T.; Nguyen, G.T.; Le, T.T.; Le, N.T.; Chang, H.C.; Pham, M.D.; Conrad, U.; Chu, H.H. Nanodiamond enhances immune responses in mice against recombinant HA/H7N9 protein. J. Nanobiotechnol. 2017, 15, 69–81. [Google Scholar] [CrossRef]
- Puzyr, A.P.; Baron, A.V.; Purtov, K.V.; Bortnikov, E.V.; Skobelev, N.N.; Mogilnaya, O.A.; Bondar, V.S. Nanodiamonds with novel properties: A biological study. Diam. Relat. Mater. 2007, 16, 2124–2128. [Google Scholar] [CrossRef]
- Wang, E.; Sandoval, R.M.; Campos, S.B.; Molitoris, B.A. Rapid diagnosis and quantification of acute kidney injury using fluorescent ratio-metric determination of glomerular filtration rate in the rat. Am. J. Physiol. Renal Physiol. 2010, 299, 1048–1055. [Google Scholar] [CrossRef]
- Li, Y.; Wu, J.; Xu, L.; Wu, Q.; Wan, Z.; Li, L.; Yu, H.; Li, X.; Li, K.; Zhang, Q.; et al. Regulation of Leukocyte Recruitment to the Spleen and Peritoneal Cavity during Pristane-Induced Inflammation. J. Immunol. Res. 2017, 2017, 9891348. [Google Scholar] [CrossRef]







| Sample | Zeta Potential (mV) | Average Size (nm) | PdI |
|---|---|---|---|
| NDA135b | −20 ± −3 | 353 ± 140 | 0.4 |
| ND | −35 ± −7 | 73 ± 28 | 0.2 |
| ND-PEI-RNA | −28 ± −4 | 120 ± 15 | 0.24 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Křivohlavá, R.; Neuhӧferová, E.; Jakobsen, K.Q.; Benson, V. Knockdown of microRNA-135b in Mammary Carcinoma by Targeted Nanodiamonds: Potentials and Pitfalls of In Vivo Applications. Nanomaterials 2019, 9, 866. https://doi.org/10.3390/nano9060866
Křivohlavá R, Neuhӧferová E, Jakobsen KQ, Benson V. Knockdown of microRNA-135b in Mammary Carcinoma by Targeted Nanodiamonds: Potentials and Pitfalls of In Vivo Applications. Nanomaterials. 2019; 9(6):866. https://doi.org/10.3390/nano9060866
Chicago/Turabian StyleKřivohlavá, Romana, Eva Neuhӧferová, Katrine Q. Jakobsen, and Veronika Benson. 2019. "Knockdown of microRNA-135b in Mammary Carcinoma by Targeted Nanodiamonds: Potentials and Pitfalls of In Vivo Applications" Nanomaterials 9, no. 6: 866. https://doi.org/10.3390/nano9060866
APA StyleKřivohlavá, R., Neuhӧferová, E., Jakobsen, K. Q., & Benson, V. (2019). Knockdown of microRNA-135b in Mammary Carcinoma by Targeted Nanodiamonds: Potentials and Pitfalls of In Vivo Applications. Nanomaterials, 9(6), 866. https://doi.org/10.3390/nano9060866