Engineering a SERS Sensing Nanoplatform with Self-Sterilization for Undifferentiated and Rapid Detection of Bacteria
Abstract
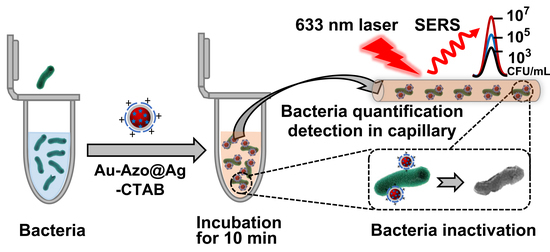
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Preparation of Au Nanoparticles (NPs)
2.3. Preparation of Au−Azo NPs
2.4. Preparation of Au−Azo@Ag NPs
2.5. Preparation of Au−Azo@Ag−CTAB NPs
2.6. Ultraviolet-Visible (UV-vis) Spectral Characterization
2.7. Dynamic Light Scattering (DLS) and Zeta Potential Characterization
2.8. Field Emission Scanning Electron Microscopy (FE-SEM) Characterization
2.9. Raman Spectroscopy Characterization
2.10. Bacterial Culture
2.11. Bacterial Quantification Using Au−Azo@Ag−CTAB NPs
2.12. Bacterial Detection and Inactivation in Tap Water
3. Results
3.1. Synthesis and Characterization of Au−Azo@Ag−CTAB NPs
3.1.1. Optimization of Synthesis Conditions of Au NPs
3.1.2. Optimization of Synthesis Conditions of Au−Azo NPs
3.1.3. Optimization of Synthesis Conditions of Au−Azo@Ag NPs
3.1.4. Optimization of Synthesis Conditions of Au−Azo@Ag−CTAB NPs
3.1.5. Characterization of Au−Azo@Ag−CTAB NPs
3.2. Ability of Au−Azo@Ag−CTAB NPs to Capture, Detect and Inactivate Bacteria
3.3. Quantitative Detection and Inactivation of Bacteria in Tap Water
4. Conclusions and Future Perspective
Abbreviation
| SERS | Surface-enhanced Raman scattering |
| Au NPs | Gold nanoparticles |
| CTAB | Cetyltrimethylammonium bromide |
| PBS | phosphate buffered saline solution |
| SEM | Scanning electron microscopy |
| POCT | Point-of-care testing |
| RSD | Relative standard deviation |
| S. aureus | Staphylococcus aureus |
| E. coli | Escherichia coli |
| CFU | Colony-forming unit |
| UV-vis | Ultraviolet-visible |
| AgNO3 | Silver nitrate |
| LOD | Limit of detection |
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Prakash, O.; Sil, S.; Verma, T.; Umapathy, S. Direct Detection of Bacteria Using Positively Charged Ag/Au Bimetallic Nanoparticles: A Label-free Surface-Enhanced Raman Scattering Study Coupled with Multivariate Analysis. J. Phys. Chem. C 2020, 124, 861–869. [Google Scholar] [CrossRef] [Green Version]
- Cao, Z.; Li, C.; Yang, X.; Wang, S.; Zhang, X.; Zhao, C.; Xue, B.; Gao, C.; Zhou, H.; Yang, Y.; et al. Rapid Quantitative Detection of Live Escherichia coli Based on Chronoamperometry. Biosensors 2022, 12, 845. [Google Scholar] [CrossRef] [PubMed]
- Al-Hindi, R.R.; Teklemariam, A.D.; Alharbi, M.G.; Alotibi, I.; Azhari, S.A.; Qadri, I.; Alamri, T.; Harakeh, S.; Applegate, B.M.; Bhunia, A.k. Bacteriophage-Based Biosensors: A Platform for Detection of Foodborne Bacterial Pathogens from Food and Environment. Biosensors 2022, 12, 905. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Sun, Y.; Li, X.; Wang, L.; Xu, Y.; He, L.; Li, G. Recent Advances in Dual Recognition Based Surface Enhanced Raman Scattering for Pathogenic Bacteria Detection: A Review. Anal. Chim. Acta 2021, 1157, 338279. [Google Scholar] [CrossRef]
- Kaur, K.; Chelangat, W.; Druzhinin, S.I.; Karuri, N.; Müller, M.; Schönherr, H. Quantitative E. coli Enzyme Detection in Reporter Hydrogel-Coated Paper Using a Smartphone Camera. Biosensors 2021, 11, 25. [Google Scholar] [CrossRef]
- Yan, C.; Sun, Y.; Yao, M.; Jin, X.; Yang, Q.; Wu, W. pH-Responsive Nanoparticles and Automated Detection Apparatus for Dual Detection of Pathogenic Bacteria. Sens. Actuators B Chem. 2022, 354, 131117. [Google Scholar] [CrossRef]
- Angelopoulou, M.; Petrou, P.; Misiakos, K.; Raptis, I.; Kakabakos, S. Simultaneous Detection of Salmonella typhimurium and Escherichia coli O157:H7 in Drinking Water and Milk with Mach-Zehnder Interferometers Monolithically Integrated on Silicon Chips. Biosensors 2022, 12, 507. [Google Scholar] [CrossRef]
- Liu, Y.; Zhu, W.; Yuan, Q.; Hu, J.; Zhang, X.; Shen, A. Photoreduced Ag+ Surrounding Single Poly(4-cyanostyrene) Nanoparticles for Undifferentiated SERS Sensing and Killing of Bacteria. Talanta 2022, 245, 123450. [Google Scholar] [CrossRef]
- Chu, H.; Huang, Y.; Zhao, Y. Silver Nanorod Arrays as a Surface-Enhanced Raman Scattering Substrate for Foodborne Pathogenic Bacteria Detection. Appl. Spectrosc. 2008, 62, 922–931. [Google Scholar] [CrossRef]
- Kang, D.K.; Ali, M.M.; Zhang, K.; Huang, S.S.; Peterson, E.; Digman, M.A.; Gratton, E.; Zhao, W. Rapid Detection of Single Bacteria in Unprocessed Blood Using Integrated Comprehensive Droplet Digital Detection. Nat. Commun. 2014, 5, 5427. [Google Scholar] [CrossRef]
- Ottesen, E.A.; Hong, J.W.; Quake, S.R.; Leadbetter, J.R. Microfluidic Digital PCR Enables Multigene Analysis of Individual Environmental Bacteria. Science 2006, 314, 1464–1467. [Google Scholar] [CrossRef] [Green Version]
- Millenbaugh, N.J.; Baskin, J.B.; DeSilva, M.N.; Elliott, W.R.; Glickman, R.D. Photothermal Killing of Staphylococcus aureus Using Antibody-Targeted Gold Nanoparticles. Int. J. Nanomed. 2015, 10, 1953–1960. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, Z.; Zhou, Z.; Yang, M.; Lin, C.; Zhao, Z.; Huang, D.; Chen, Z.; Gao, J. Multifunctional Ag@Fe2O3 Yolk-Shell Nanoparticles for Simultaneous Capture, Kill, and Removal of Pathogen. J. Mater. Chem. 2011, 21, 16344. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Z.; Xiao, R.; Cheng, S.; Wang, S.; Shi, L.; Wang, C.; Qi, K.; Wang, S. A Universal SERS-Label Immunoassay for Pathogen Bacteria Detection Based on Fe3O4@Au-Aptamer Separation and Antibody-Protein A Orientation Recognition. Anal. Chim. Acta 2021, 1160, 338421. [Google Scholar] [CrossRef] [PubMed]
- Mosier-Boss, P.A. Review on SERS of Bacteria. Biosensors 2017, 7, 51. [Google Scholar] [CrossRef] [Green Version]
- Laing, S.; Gracie, K.; Faulds, K. Multiplex In Vitro Detection Using SERS. Chem. Soc. Rev. 2016, 45, 1901–1918. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Wang, C.; Xiao, R.; Tang, L.; Huang, J.; Wu, D.; Liu, S.; Wang, Y.; Zhang, D.; Wang, S.; et al. Sensitive and Specific Detection of Clinical Bacteria via Vancomycin-Modified Fe3O4@Au Nanoparticles and Aptamer-Functionalized SERS Tags. J. Mater. Chem. B 2018, 6, 3751–3761. [Google Scholar] [CrossRef]
- Xiang, X.; Feng, S.; Chen, J.; Feng, J.; Hou, Y.; Ruan, Y.; Weng, X.; Milcovich, G. Gold Nanoparticles/Electrochemically Expanded Graphite Composite: A Bifunctional Platform Toward Glucose Sensing and SERS Applications. J. Electroanal. Chem. 2019, 851, 113471. [Google Scholar] [CrossRef]
- Zong, S.; Wang, Z.; Chen, H.; Yang, J.; Cui, Y. Surface Enhanced Raman Scattering Traceable and Glutathione Responsive Nanocarrier for the Intracellular Drug Delivery. Anal. Chem. 2013, 85, 2223–2230. [Google Scholar] [CrossRef]
- Hu, Z.; Zhou, X.; Duan, J.; Wu, X.; Wu, J.; Zhang, P.; Liang, W.; Guo, J.; Cai, H.; Sun, P.; et al. Aptamer-Based Novel Ag-Coated Magnetic Recognition and SERS Nanotags with Interior Nanogap Biosensor for Ultrasensitive Detection of Protein Biomarker. Sens. Actuators B Chem. 2021, 334, 129640. [Google Scholar] [CrossRef]
- Lin, S.; Hasi, W.; Lin, X.; Han, S.; Xiang, T.; Liang, S.; Wang, L. Lab-On-Capillary Platform for On-Site Quantitative SERS Analysis of Surface Contaminants Based on Au@4-MBA@Ag Core–Shell Nanorods. ACS Sens. 2020, 5, 1465–1473. [Google Scholar] [CrossRef]
- Wang, C.; Wang, J.; Li, M.; Qu, X.; Zhang, K.; Rong, Z.; Xiao, R.; Wang, S. A Rapid SERS Method for Label-Free Bacteria Detection Using Polyethylenimine-Modified Au-Coated Magnetic Microspheres and Au@Ag Nanoparticles. Analyst. 2016, 141, 6226–6238. [Google Scholar] [CrossRef]
- Gao, X.; Yin, Y.; Wu, H.; Hao, Z.; Li, J.; Wang, S.; Liu, Y. Integrated SERS Platform for Reliable Detection and Photothermal Elimination of Bacteria in Whole Blood Samples. Anal. Chem. 2021, 93, 1569–1577. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Zhang, Z.; Chen, L.; Lin, Y.; Zeng, R.; Xu, J.; Chen, S.; Zhang, J.; Cai, H.; Zhou, H.; et al. Multifunctional Au Nano-Bridged Nanogap Probes as ICP-MS/SERS Dual-Signal Tags and Signal Amplifiers for Bacteria Discriminating, Quantitative Detecting and Photothermal Bactericidal Activity. Biosens. Bioelectron. 2022, 212, 114414. [Google Scholar] [CrossRef] [PubMed]
- Cheng, D.; Yu, M.; Fu, F.; Han, W.; Li, G.; Xie, J.; Song, Y.; Swihart, M.; Song, E.; Wang, S.; et al. Dual Recognition Strategy for Specific and Sensitive Detection of Bacteria Using Aptamer-Coated Magnetic Beads and Antibiotic-Capped Gold Nanoclusters. Anal. Chem. 2015, 88, 820–825. [Google Scholar] [CrossRef]
- Luo, K.; Ryu, J.; Seol, I.H.; Jeong, K.B.; You, S.M.; Kim, Y.R. Paper-Based Radial Chromatographic Immunoassay for the Detection of Pathogenic Bacteria in Milk. ACS Appl. Mater. Interfaces 2019, 11, 46472–46478. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Tang, Y.; Chu, K.; Gao, T.; Smith, Z.J. High-Resolution Low-Power Hyperspectral Line-Scan Imaging of Fast Cellular Dynamics Using Azo-Enhanced Raman Scattering Probes. J. Am. Chem. Soc. 2022, 144, 15314–15323. [Google Scholar] [CrossRef]
- Tang, Y.; Zhuang, Y.; Zhang, S.; Smith, Z.; Li, Y.; Mu, X.; Li, M.; He, C.; Zheng, X.; Pan, F.; et al. Azo-Enhanced Raman Scattering for Enhancing the Sensitivity and Tuning the Frequency of Molecular Vibrations. ACS Cent. Sci. 2021, 7, 768–780. [Google Scholar] [CrossRef]
- Hayden, S.C.; Zhao, G.; Saha, K.; Phillips, R.L.; Li, X.; Miranda, O.R.; Rotello, V.M.; El-Sayed, M.A.; Schmidt-Krey, I.; Bunz, U.H.F. Aggregation and Interaction of Cationic Nanoparticles on Bacterial Surfaces. J. Am. Chem. Soc. 2012, 134, 6920–6923. [Google Scholar] [CrossRef] [PubMed]
- Hussain, S.; Lv, F.; Qi, R.; Senthilkumar, T.; Zhao, H.; Chen, Y.; Liu, L.; Wang, S. Förster Resonance Energy Transfer Mediated Rapid and Synergistic Discrimination of Bacteria over Fungi Using a Cationic Conjugated Glycopolymer. ACS Appl. Bio Mater. 2020, 3, 20–28. [Google Scholar] [CrossRef]
- Fernández-Grajera, M.; Gallardo-Moreno, A.M.; Luque-Agudo, V.; González-Martín, M.L.; Hierro-Oliva, M. Bacterial Response to the Surface Aging of PLA Matrices Loaded with Active Compounds. Polymers 2022, 14, 4976. [Google Scholar] [CrossRef] [PubMed]
- Abduraimova, A.; Molkenova, A.; Duisembekova, A.; Mulikova, T.; Kanayeva, D.; Atabaev, T.S. Cetyltrimethylammonium Bromide (CTAB)-Loaded SiO2-Ag Mesoporous Nanocomposite as an Efficient Antibacterial Agent. Nanomaterials 2021, 11, 477. [Google Scholar] [CrossRef] [PubMed]
- Daniel, M.; Astruc, D. Gold Nanoparticles: Assembly, Supramolecular Chemistry, Quantum-Size-Related Properties, and Applications toward Biology, Catalysis, and Nanotechnology. Chem. Rev. 2004, 104, 293–346. [Google Scholar] [CrossRef] [PubMed]
- Deeb, C.; Zhou, X.; Plain, J.; Wiederrecht, G.P.; Bachelot, R.; Russell, M.; Jain, P.K. Size Dependence of the Plasmonic Near-Field Measured via Single-Nanoparticle Photoimaging. J. Phys. Chem. C 2013, 117, 10669–10676. [Google Scholar] [CrossRef]
- Bi, L.; Wang, X.; Cao, X.; Liu, L.; Bai, C.; Zheng, Q.; Choo, J.; Chen, L. SERS-Active Au@Ag Core-Shell Nanorod (Au@AgNR) Tags for Ultrasensitive Bacteria Dtetion and Antibiotic-Susceptibility Testing. Talanta 2020, 220, 121397. [Google Scholar] [CrossRef]





| Sample | Culturing Result (CFU/mL) | Detected Result (CFU/mL) | Relative Recovery (%) | RSD 1 (%) |
|---|---|---|---|---|
| E. coli−1 | 2.40 × 103 | 2.59 × 103 | 108 | 4.2 |
| E. coli−2 | 3.50 × 103 | 3.85 × 103 | 110 | 4.5 |
| E. coli−3 | 4.85 × 104 | 5.24 × 104 | 108 | 4.1 |
| S. aureus−1 | 5.70 × 103 | 6.10 × 103 | 107 | 4.3 |
| S. aureus−2 | 7.50 × 103 | 7.95 × 103 | 106 | 4.2 |
| S. aureus−3 | 3.81 × 104 | 4.04 × 104 | 106 | 4.3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, J.; Zhu, W.; Zhou, J.; Zhao, B.-C.; Pan, Y.-Y.; Ye, Y.; Shen, A.-G. Engineering a SERS Sensing Nanoplatform with Self-Sterilization for Undifferentiated and Rapid Detection of Bacteria. Biosensors 2023, 13, 75. https://doi.org/10.3390/bios13010075
Cao J, Zhu W, Zhou J, Zhao B-C, Pan Y-Y, Ye Y, Shen A-G. Engineering a SERS Sensing Nanoplatform with Self-Sterilization for Undifferentiated and Rapid Detection of Bacteria. Biosensors. 2023; 13(1):75. https://doi.org/10.3390/bios13010075
Chicago/Turabian StyleCao, Jun, Wei Zhu, Ji Zhou, Bai-Chuan Zhao, Yao-Yu Pan, Yong Ye, and Ai-Guo Shen. 2023. "Engineering a SERS Sensing Nanoplatform with Self-Sterilization for Undifferentiated and Rapid Detection of Bacteria" Biosensors 13, no. 1: 75. https://doi.org/10.3390/bios13010075
APA StyleCao, J., Zhu, W., Zhou, J., Zhao, B. -C., Pan, Y. -Y., Ye, Y., & Shen, A. -G. (2023). Engineering a SERS Sensing Nanoplatform with Self-Sterilization for Undifferentiated and Rapid Detection of Bacteria. Biosensors, 13(1), 75. https://doi.org/10.3390/bios13010075