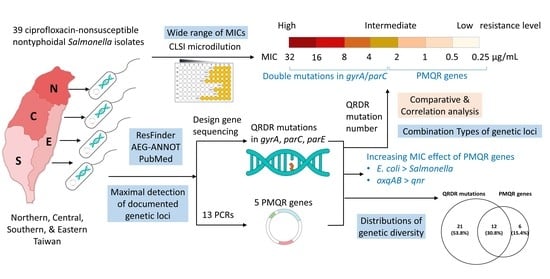
Genotypic Diversity of Ciprofloxacin Nonsusceptibility and Its Relationship with Minimum Inhibitory Concentrations in Nontyphoidal Salmonella Clinical Isolates in Taiwan
Abstract
:1. Introduction
2. Results
2.1. Concomitant Resistance to Ampicillin and Ceftriaxone in the CIP-Nonsusceptible NTS Isolates
2.2. Detected Genomic Point Mutations in Three QRDR Genes
2.3. Detected Five PMQR Genes
2.4. Distribution of Detected QRDR Mutations and PMQR Genes
2.5. Relationship between Genetic Mechanisms and the MIC of CIP
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Serotyping
4.2. Antibiotic Susceptibility Test
4.3. Searching Mutations and Genes Associated with Quinolone Resistance in Three Databases
4.4. Sequencing for the Detection of Genomic Mutations in the QRDR
4.5. PCRs for the Detection of the 13 PMRQ Genes
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Su, L.-H.; Chiu, C.-H.; Chu, C.; Ou, J.T. Antimicrobial Resistance in Nontyphoid Salmonella Serotypes: A Global Challenge. Clin. Infect. Dis. 2004, 39, 546–551. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Campos, M.J.; Palomo, G.; Hormeño, L.; Herrera-León, S.; Domínguez, L.; Vadillo, S.; Píriz, S.; Quesada, A. Prevalence of quinolone resistance determinants in non-typhoidal Salmonella isolates from human origin in Extremadura, Spain. Diagn. Microbiol. Infect. Dis. 2014, 79, 64–69. [Google Scholar] [CrossRef]
- Ceyssens, P.-J.; Mattheus, W.; Vanhoof, R.; Bertrand, S. Trends in Serotype Distribution and Antimicrobial Susceptibility in Salmonella enterica Isolates from Humans in Belgium, 2009 to 2013. Antimicrob. Agents Chemother. 2015, 59, 544–552. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.-L.; Lu, M.-C.; Shao, P.-L.; Lu, P.-L.; Chen, Y.-H.; Cheng, S.-H.; Ko, W.-C.; Lin, C.-Y.; Wu, T.-S.; Yen, M.-Y.; et al. Nationwide surveillance of antimicrobial resistance among clinically important Gram-negative bacteria, with an emphasis on carbapenems and colistin: Results from the Surveillance of Multicenter Antimicrobial Resistance in Taiwan (SMART) in 2018. Int. J. Antimicrob. Agents 2019, 54, 318–328. [Google Scholar] [CrossRef] [PubMed]
- Medalla, F.; Gu, W.; Friedman, C.R.; Judd, M.; Folster, J.; Griffin, P.M.; Hoekstra, R.M. Increased Incidence of Antimicrobial-Resistant Nontyphoidal Salmonella Infections, United States, 2004–2016. Emerg. Infect. Dis. 2021, 27, 1662–1672. [Google Scholar] [CrossRef]
- Medalla, F.; Hoekstra, R.M.; Whichard, J.M.; Barzilay, E.J.; Chiller, T.M.; Joyce, K.; Rickert, R.; Krueger, A.; Stuart, A.; Griffin, P.M. Increase in Resistance to Ceftriaxone and Nonsusceptibility to Ciprofloxacin and Decrease in Multidrug Resistance AmongSalmonellaStrains, United States, 1996–2009. Foodborne Pathog. Dis. 2013, 10, 302–309. [Google Scholar] [CrossRef] [PubMed]
- Tadesse, G.; Tessema, T.S.; Beyene, G.; Aseffa, A. Molecular epidemiology of fluoroquinolone resistant Salmonella in Africa: A systematic review and meta-analysis. PLoS ONE 2018, 13, e0192575. [Google Scholar] [CrossRef] [Green Version]
- Redgrave, L.; Sutton, S.B.; Webber, M.; Piddock, L.J. Fluoroquinolone resistance: Mechanisms, impact on bacteria, and role in evolutionary success. Trends Microbiol. 2014, 22, 438–445. [Google Scholar] [CrossRef]
- Cuypers, W.; Jacobs, J.; Wong, V.; Klemm, E.J.; Deborggraeve, S.; Van Puyvelde, S. Fluoroquinolone resistance in Salmonella: Insights by whole-genome sequencing. Microb. Genom. 2018, 4, e000195. [Google Scholar] [CrossRef]
- Correia, S.; Poeta, P.; Hebraud, M.; Capelo, J.L.; Igrejas, G. Mechanisms of quinolone action and resistance: Where do we stand? J. Med. Microbiol. 2017, 66, 551–559. [Google Scholar] [CrossRef] [PubMed]
- Kao, C.-Y.; Chen, C.-A.; Liu, Y.-F.; Wu, H.-M.; Chiou, C.-S.; Yan, J.-J.; Wu, J.-J. Molecular characterization of antimicrobial susceptibility of Salmonella isolates: First identification of a plasmid carrying qnrD or oqxAB in Taiwan. J. Microbiol. Immunol. Infect. 2017, 50, 214–223. [Google Scholar] [CrossRef] [Green Version]
- Abgottspon, H.; Zurfluh, K.; Nüesch-Inderbinen, M.; Hächler, H.; Stephan, R. Quinolone Resistance Mechanisms in Salmonella enterica Serovars Hadar, Kentucky, Virchow, Schwarzengrund, and 4,5,12:i:−, Isolated from Humans in Switzerland, and Identification of a NovelqnrDVariant, qnrD2, inS. Hadar. Antimicrob. Agents Chemother. 2014, 58, 3560–3563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Acheampong, G.; Owusu, M.; Owusu-Ofori, A.; Osei, I.; Sarpong, N.; Sylverken, A.; Kung, H.-J.; Cho, S.-T.; Kuo, C.-H.; Park, S.E.; et al. Chromosomal and plasmid-mediated fluoroquinolone resistance in human Salmonella enterica infection in Ghana. BMC Infect. Dis. 2019, 19, 898. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karp, B.E.; Campbell, D.; Chen, J.C.; Folster, J.P.; Friedman, C.R. Plasmid-mediated quinolone resistance in human non-typhoidal Salmonella infections: An emerging public health problem in the United States. Zoonoses Public Health 2018, 65, 838–849. [Google Scholar] [CrossRef]
- Kim, S.-Y.; Lee, S.-K.; Park, M.-S.; Na, H.-T. Analysis of the Fluoroquinolone Antibiotic Resistance Mechanism of Salmonella enterica Isolates. J. Microbiol. Biotechnol. 2016, 26, 1605–1612. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.; Park, N.; Yun, S.; Hur, E.; Song, J.; Lee, H.; Kim, Y.; Ryu, S. Presence of plasmid-mediated quinolone resistance (PMQR) genes in non-typhoidal Salmonella strains with reduced susceptibility to fluoroquinolones isolated from human salmonellosis in Gyeonggi-do, South Korea from 2016 to 2019. Gut Pathog. 2021, 13, 35. [Google Scholar] [CrossRef]
- Crump, J.A.; Barrett, T.J.; Nelson, J.T.; Angulo, F.J. Reevaluating Fluoroquinolone Breakpoints for Salmonella enterica Serotype Typhi and for Non-Typhi Salmonellae. Clin. Infect. Dis. 2003, 37, 75–81. [Google Scholar] [CrossRef]
- Sjölund-Karlsson, M.; Howie, R.; Crump, J.A.; Whichard, J.M. Fluoroquinolone Susceptibility Testing of Salmonella enterica: Detection of Acquired Resistance and Selection of Zone Diameter Breakpoints for Levofloxacin and Ofloxacin. J. Clin. Microbiol. 2014, 52, 877–884. [Google Scholar] [CrossRef] [Green Version]
- Humphries, R.M.; Fang, F.; Aarestrup, F.; Hindler, J.A. In Vitro Susceptibility Testing of Fluoroquinolone Activity Against Salmonella: Recent Changes to CLSI Standards. Clin. Infect. Dis. 2012, 55, 1107–1113. [Google Scholar] [CrossRef] [Green Version]
- Rodríguez-Martínez, J.-M.; López-Cerero, L.; Díaz-De-Alba, P.; Chamizo-López, F.J.; Polo-Padillo, J.; Pascual, A. Assessment of a phenotypic algorithm to detect plasmid-mediated quinolone resistance in Enterobacteriaceae. J. Antimicrob. Chemother. 2015, 71, 845–847. [Google Scholar] [CrossRef] [Green Version]
- Eguale, T.; Birungi, J.; Asrat, D.; Njahira, M.N.; Njuguna, J.; Gebreyes, W.A.; Gunn, J.S.; Djikeng, A.; Engidawork, E. Genetic markers associated with resistance to beta-lactam and quinolone antimicrobials in non-typhoidal Salmonella isolates from humans and animals in central Ethiopia. Antimicrob. Resist. Infect. Control. 2017, 6, 13. [Google Scholar] [CrossRef] [Green Version]
- Cebríán, L.; Escribano, I.; Rodríguez, J.; Royo, G. Alterations in thegyrA andparC Genes inSalmonellaspp. FollowingIn VitroExposure to Fluoroquinolones. J. Chemother. 2006, 18, 250–254. [Google Scholar] [CrossRef]
- Jeong, H.S.; Bae, I.K.; Shin, J.H.; Jung, H.J.; Kim, S.H.; Lee, J.Y.; Oh, S.H.; Kim, H.R.; Chang, C.L.; Kho, W.-G.; et al. Prevalence of Plasmid-mediated Quinolone Resistance and Its Association with Extended-spectrum Beta-lactamase and AmpC Beta-lactamase in Enterobacteriaceae. Ann. Lab. Med. 2011, 31, 257–264. [Google Scholar] [CrossRef]
- Yan, J.-J.; Chiou, C.-S.; Lauderdale, T.-L.Y.; Tsai, S.-H.; Wu, J.-J. Cephalosporin and Ciprofloxacin Resistance inSalmonella, Taiwan. Emerg. Infect. Dis. 2005, 18, 947–950. [Google Scholar] [CrossRef] [PubMed]
- Gunell, M.; Webber, M.; Kotilainen, P.; Lilly, A.J.; Caddick, J.M.; Jalava, J.; Huovinen, P.; Siitonen, A.; Hakanen, A.J.; Piddock, L. Mechanisms of Resistance in Nontyphoidal Salmonella enterica Strains Exhibiting a Nonclassical Quinolone Resistance Phenotype. Antimicrob. Agents Chemother. 2009, 53, 3832–3836. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ling, J.M.; Chan, E.W.; Lam, A.W.; Cheng, A.F. Mutations in Topoisomerase Genes of Fluoroquinolone-Resistant Salmonellae in Hong Kong. Antimicrob. Agents Chemother. 2003, 47, 3567–3573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, M.-X.; Zhang, J.-F.; Sun, Y.-H.; Li, R.-S.; Lin, X.-L.; Yang, L.; Webber, M.A.; Jiang, H.-X. Contribution of Different Mechanisms to Ciprofloxacin Resistance in Salmonella spp. Front. Microbiol. 2021, 12, 663731. [Google Scholar] [CrossRef] [PubMed]
- Năşcuţiu, A.-M. The tip of the iceberg: Quinolone-resistance conferred by mutations in gyrA gene in non-typhoidal Salmonella strains. Roum. Arch. Microbiol. Immunol. 2012, 71, 17–23. [Google Scholar]
- Hopkins, K.L.; Davies, R.H.; Threlfall, E.J. Mechanisms of quinolone resistance in Escherichia coli and Salmonella: Recent developments. Int. J. Antimicrob. Agents 2005, 25, 358–373. [Google Scholar] [CrossRef]
- Hooper, D.C.; Jacoby, G.A. Mechanisms of drug resistance: Quinolone resistance. Ann. N. Y. Acad. Sci. 2015, 1354, 12–31. [Google Scholar] [CrossRef] [Green Version]
- Lian, X.; Wang, X.; Liu, X.; Xia, J.; Fang, L.; Sun, J.; Liao, X.; Liu, Y. oqxAB-Positive IncHI2 Plasmid pHXY0908 Increase Salmonella enterica Serotype Typhimurium Strains Tolerance to Ciprofloxacin. Front. Cell. Infect. Microbiol. 2019, 9, 242. [Google Scholar] [CrossRef] [Green Version]
- Wong, M.H.-Y.; Chan, E.W.; Liu, L.Z.; Chen, S. PMQR genes oqxAB and aac(6â€2)Ib-cr accelerate the development of fluoroquinolone resistance in Salmonella typhimurium. Front. Microbiol. 2014, 5, 521. [Google Scholar] [CrossRef] [PubMed]
- Robicsek, A.; Strahilevitz, J.; Jacoby, G.A.; Macielag, M.; Abbanat, D.; Park, C.H.; Bush, K.; Hooper, D.C. Fluoroquinolone-modifying enzyme: A new adaptation of a common aminoglycoside acetyltransferase. Nat. Med. 2006, 12, 83–88. [Google Scholar] [CrossRef] [PubMed]
- Hopkins, K.L.; Wootton, L.; Day, M.R.; Threlfall, E.J. Plasmid-mediated quinolone resistance determinant qnrS1 found in Salmonella enterica strains isolated in the UK. J. Antimicrob. Chemother. 2007, 59, 1071–1075. [Google Scholar] [CrossRef] [Green Version]
- Chen, K.; Yang, C.; Dong, N.; Xie, M.; Ye, L.; Chan, E.W.C.; Chen, S. Evolution of Ciprofloxacin Resistance-Encoding Genetic Elements in Salmonella. mSystems 2020, 5, e01234-20. [Google Scholar] [CrossRef] [PubMed]
- Monte, D.F.; Nethery, M.A.; Barrangou, R.; Landgraf, M.; Fedorka-Cray, P.J. Whole-genome sequencing analysis and CRISPR genotyping of rare antibiotic-resistant Salmonella enterica serovars isolated from food and related sources. Food Microbiol. 2021, 93, 103601. [Google Scholar] [CrossRef] [PubMed]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing, 30th ed.; CLSI supplement M100; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2020. [Google Scholar]
- Reyna, F.; Huesca, M.; Gonzalez, V.; Fuchs, L.Y. Salmonella typhimurium gyrA mutations associated with fluoroquinolone resistance. Antimicrob. Agents Chemother. 1995, 39, 1621–1623. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turner, A.K.; Nair, S.; Wain, J. The acquisition of full fluoroquinolone resistance in Salmonella Typhi by accumulation of point mutations in the topoisomerase targets. J. Antimicrob. Chemother. 2006, 58, 733–740. [Google Scholar] [CrossRef] [Green Version]
- Barnard, F.M.; Maxwell, A. Interaction between DNA gyrase and quinolones: Effects of alanine mutations at GyrA subunit residues Ser(83) and Asp(87). Antimicrob. Agents Chemother. 2001, 45, 1994–2000. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cesaro, A.; Bettoni, R.R.; Lascols, C.; Merens, A.; Soussy, C.J.; Cambau, E. Low selection of topoisomerase mutants from strains of Escherichia coli harbouring plasmid-borne qnr genes. J. Antimicrob. Chemother. 2008, 61, 1007–1015. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.C.; Chen, T.H.; Wang, Y.C.; Chang, C.C.; Hsuan, S.L.; Chang, Y.C.; Yeh, K.S. Analysis of ciprofloxacin-resistant Salmonella strains from swine, chicken, and their carcasses in Taiwan and detection of parC resistance mutations by a mismatch amplification mutation assay PCR. J. Food Prot. 2009, 72, 14–20. [Google Scholar] [CrossRef] [PubMed]
- Yonezawa, M.; Takahata, M.; Matsubara, N.; Watanabe, Y.; Narita, H. DNA gyrase gyrA mutations in quinolone-resistant clinical isolates of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 1995, 39, 1970–1972. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al-Emran, H.M.; Heisig, A.; Dekker, D.; Adu-Sarkodie, Y.; Cruz Espinoza, L.M.; Panzner, U.; von Kalckreuth, V.; Marks, F.; Park, S.E.; Sarpong, N.; et al. Detection of a Novel gyrB Mutation Associated With Fluoroquinolone-Nonsusceptible Salmonella enterica serovar Typhimurium Isolated From a Bloodstream Infection in Ghana. Clin. Infect. Dis. 2016, 62 (Suppl. 1), S47–S49. [Google Scholar] [CrossRef] [Green Version]
- Eaves, D.J.; Randall, L.; Gray, D.T.; Buckley, A.; Woodward, M.J.; White, A.P.; Piddock, L.J. Prevalence of mutations within the quinolone resistance-determining region of gyrA, gyrB, parC, and parE and association with antibiotic resistance in quinolone-resistant Salmonella enterica. Antimicrob. Agents Chemother. 2004, 48, 4012–4015. [Google Scholar] [CrossRef] [Green Version]
- Saenz, Y.; Zarazaga, M.; Brinas, L.; Ruiz-Larrea, F.; Torres, C. Mutations in gyrA and parC genes in nalidixic acid-resistant Escherichia coli strains from food products, humans and animals. J. Antimicrob. Chemother. 2003, 51, 1001–1005. [Google Scholar] [CrossRef]
- O’Regan, E.; Quinn, T.; Pages, J.M.; McCusker, M.; Piddock, L.; Fanning, S. Multiple regulatory pathways associated with high-level ciprofloxacin and multidrug resistance in Salmonella enterica serovar enteritidis: Involvement of RamA and other global regulators. Antimicrob. Agents Chemother. 2009, 53, 1080–1087. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Isolate ID | QRDR Mutations | PMQR Genes | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| gyrA | parC | parE | aac(6′)-Ib-cr | oqxA | oqxB | qnrB | qnrS | ||||||||||
| C248T (Ser83Phe) | C248A (Ser83Tyr) | G259A (Asp87Asn) | A260G (Asp87Gly) | C170G (Thr57Ser) | A238C (Ser80Arg) | G239T (Ser80IIe) | G250A (Glu84Lys) | T1372C (Ser458Pro) | |||||||||
| C01 | + | – | – | + | + | + | – | – | + | – | – | – | – | – | |||
| C02 | + | – | – | + | + | + | – | – | - | – | – | – | – | – | |||
| C03 | + | – | – | + | + | + | – | – | + | – | – | – | – | – | |||
| C04 | + | – | – | + | + | + | – | – | + | – | – | – | – | – | |||
| C05 | + | – | + | – | + | – | – | + | – | – | – | – | – | – | |||
| C06 | + | – | – | + | + | + | – | – | + | – | – | – | – | – | |||
| C07 | + | – | – | + | + | + | – | – | + | – | – | – | – | – | |||
| C08 | – | + | – | – | – | – | – | – | – | – | + | + | – | – | |||
| C09 | – | + | – | – | – | – | – | – | – | – | + | + | – | – | |||
| C10 | – | + | – | – | – | – | – | – | – | – | + | + | – | – | |||
| C11 | + | – | + | – | + | – | + | – | – | – | – | – | – | – | |||
| C12 | + | – | + | – | + | – | + | – | – | – | – | – | – | – | |||
| C13 | – | + | – | – | – | – | – | – | – | – | + | + | – | – | |||
| C14 | – | + | – | – | – | – | – | – | – | – | + | + | – | – | |||
| C15 | + | – | + | – | + | – | + | – | – | – | – | – | – | – | |||
| C16 | + | – | – | + | + | + | – | – | + | – | – | – | – | – | |||
| C17 | – | + | – | – | – | – | – | – | – | – | + | + | – | – | |||
| C18 | + | – | – | + | + | + | – | – | + | – | – | – | – | – | |||
| C19 | + | – | + | – | + | – | + | – | – | – | – | – | – | – | |||
| C20 | – | + | – | – | – | – | – | – | – | – | + | + | – | – | |||
| C21 | + | – | – | + | + | + | – | – | + | – | – | – | – | – | |||
| C22 | + | – | + | – | + | – | + | – | – | – | – | – | – | – | |||
| C23 | – | + | – | – | – | – | – | – | – | – | + | + | – | – | |||
| C24 | + | – | + | – | + | – | + | – | – | – | – | – | – | – | |||
| C25 | + | – | + | – | + | – | + | – | – | – | – | – | – | – | |||
| C26 | – | – | – | – | – | – | – | – | – | – | + | + | – | + | |||
| C27 | – | – | – | – | – | – | – | – | – | – | + | + | – | + | |||
| C28 | + | – | + | – | + | – | + | – | – | – | – | – | – | – | |||
| C29 | – | – | + | – | – | – | – | – | – | + | + | + | – | – | |||
| C30 | – | – | – | – | – | – | – | – | – | + | – | – | – | + | |||
| C31 | + | – | – | – | + | – | – | – | – | – | – | – | – | – | |||
| C32 | – | – | – | – | – | – | – | – | – | + | – | – | – | + | |||
| C33 | – | + | – | – | + | – | – | – | – | – | – | – | – | + | |||
| C34 | – | – | – | – | – | – | – | – | – | – | – | – | – | + | |||
| C35 | – | + | – | – | + | – | – | – | – | – | – | – | – | + | |||
| C36 | – | – | – | – | – | – | – | – | – | + | – | – | + | – | |||
| C37 | + | – | – | – | + | – | – | – | – | – | – | – | – | – | |||
| C38 | – | – | – | – | + | – | – | – | – | – | – | – | + | – | |||
| C39 | + | – | – | – | – | – | – | – | – | – | – | – | – | – | |||
| Total: 39 | 21 | 10 | 10 | 9 | 23 | 9 | 8 | 1 | 8 | 4 | 11 | 11 | 2 | 7 | |||
| Percentage | 53.8% | 25.6% | 25.6% | 23.0% | 58.9% | 23.0% | 20.5% | 2.6% | 20.5% | 10.3% | 28.2% | 28.2% | 5.1% | 18.0% | |||
| Group (Isolate No.) | MIC (μg/mL)/Isolate ID | Combination Type | QRDR Mutations | PMQR Genes | ||
|---|---|---|---|---|---|---|
| gyrA | parC | parE | ||||
| 1 (n = 8) | 32/C01, C03, C04, C06, C07, C16, C18, C21 | I | Ser83Phe Asp87Gly | Thr57Ser Ser80Arg | Ser458Pro | – |
| 2 (n = 10) | 16/C12, C15, C28 8/C19, C22, C24, C25 4/C11 | II | Ser83Phe Asp87Asn | Thr57Ser Ser80IIe | – | – |
| 8/C05 | III | Ser83Phe Asp87Asn | Thr57Ser Glu84Lys | – | – | |
| 8/C02 | IV | Ser83Phe Asp87Gly | Thr57Ser Ser80Arg | – | – | |
| 3 (n = 14) | 2/C29 | V | Asp87Asn | – | – | aac(6′)-Ib-cr oqxA, oqxB |
| 2/C26, C27 | VI | – | – | – | oqxA, oqxB, qnrS | |
| 1/C33, C35 | VII | Ser83Tyr | Thr57Ser | – | qnrS | |
| 1/C08, C09, C10, C13, C14, C17, C20, C23 | VIII | Ser83Tyr | – | – | oqxA, oqxB | |
| 1/C34 | IX | – | – | – | qnrS | |
| 4 (n = 7) | 0.5/C31, C37 | X | Ser83Phe | Thr57Ser | – | – |
| 0.5/36 | XI | – | – | – | aac(6′)-Ib-cr qnrB | |
| 0.5/C32 0.25/C30 | XII | – | – | – | aac(6′)-Ib-cr qnrS | |
| 0.25/C39 | XIII | Ser83Phe | – | – | – | |
| 0.25/C38 | XIV | – | Thr57Ser | – | qnrB | |
| Grouping by MICs | No. of Isolates | Number of Different MICs (μg/mL) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | 32 | ||
| 1 (32 μg/mL) | 8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 8 |
| 2 (4–16 μg/mL) | 10 | 0 | 0 | 0 | 0 | 1 | 6 | 3 | 0 |
| 3 (1–2 μg/mL) | 14 | 0 | 0 | 11 | 3 | 0 | 0 | 0 | 0 |
| 4 (0.25–0.5 μg/mL) | 7 | 3 | 4 | 0 | 0 | 0 | 0 | 0 | 0 |
| Total | 39 | 3 | 4 | 11 | 3 | 1 | 6 | 3 | 8 |
| Grouping by QRDR Mutation No. | Groups (MICs) | |||
|---|---|---|---|---|
| 1 (32 μg/mL) | 2 (4–16 μg/mL) | 3 (1–2 μg/mL) | 4 (0.25–0.5 μg/mL) | |
| 1 (5 mutations) | ||||
| Case No. (%) | 8 (100) * | 0 (0) | 0 (0) | 0 (0) |
| 2 (4 mutations) | ||||
| Case No. (%) | 0 (0) | 10 (100) * | 0 (0) | 0 (0) |
| 3 (0–3 mutations) | ||||
| Case No. (%) | 0 (0) | 0 (0) | 14 (100) * | 7 (100) * |
| Total Case No. | 8 | 10 | 14 | 7 |
| Isolate ID | Year | Region | Serotype | Disc Inhibition Test | MIC (μg/mL) | ||||
|---|---|---|---|---|---|---|---|---|---|
| CIP * | AMP † | CRO ‡ | CIP * | AMP † | CRO ‡ | ||||
| C01 | 1998 | S | – | R | R | S | 32 | >16 | ≤1 |
| C02 | 1998 | C | Schwarzengrund | R | R | S | 8 | >16 | ≤1 |
| C03 | 1998 | C | Schwarzengrund | R | R | S | 32 | >16 | ≤1 |
| C04 | 1998 | S | – | R | R | S | 32 | >16 | ≤1 |
| C05 | 1998 | S | – | R | S | S | 8 | ≤4 | ≤1 |
| C06 | 1998 | S | – | R | S | S | 32 | ≤4 | ≤1 |
| C07 | 2000 | C | Schwarzengrund | R | R | S | 32 | >16 | ≤1 |
| C08 | 2000 | E | – | R | R | S | 1 | >16 | ≤1 |
| C09 | 2000 | E | – | R | R | S | 1 | >16 | ≤1 |
| C10 | 2000 | E | – | R | R | S | 1 | >16 | ≤1 |
| C11 | 2000 | E | – | R | R | S | 8 | >16 | ≤1 |
| C12 | 2000 | E | – | R | R | S | 16 | >16 | ≤1 |
| C13 | 2000 | N | – | R | R | S | 1 | >16 | ≤1 |
| C14 | 2000 | C | Typhimurium | I | R | S | 1 | >16 | ≤1 |
| C15 | 2000 | S | – | R | R | S | 16 | >16 | ≤1 |
| C16 | 2002 | S | – | R | R | S | 32 | >16 | ≤1 |
| C17 | 2002 | S | – | R | S | S | 1 | ≤4 | 1 |
| C18 | 2002 | N | – | R | R | R | 32 | >16 | 8 |
| C19 | 2002 | E | – | R | R | S | 32 | >16 | ≤1 |
| C20 | 2002 | E | – | R | R | S | 1 | >16 | ≤1 |
| C21 | 2002 | E | – | R | R | S | 32 | >16 | ≤1 |
| C22 | 2002 | E | – | R | R | S | 8 | >16 | ≤1 |
| C23 | 2002 | C | – | R | S | S | 1 | ≤4 | ≤1 |
| C24 | 2002 | N | – | R | R | S | 8 | >16 | ≤1 |
| C25 | 2002 | C | Choleraesuis | R | R | S | 8 | >16 | ≤1 |
| C26 | 2010 | S | – | R | S | S | 2 | ≤4 | ≤1 |
| C27 | 2012 | E | – | R | R | S | 2 | >16 | ≤1 |
| C28 | 2012 | S | – | R | R | S | 16 | >16 | ≤1 |
| C29 | 2012 | C | Typhimurium | R | R | S | 2 | >16 | ≤1 |
| C30 | 2012 | C | Typhimurium | I | R | S | 0.25 | >16 | ≤1 |
| C31 | 2012 | C | – | I | R | S | 0.5 | >16 | ≤1 |
| C32 | 2012 | C | – | I | R | S | 0.5 | >16 | ≤1 |
| C33 | 2014 | C | Enteritidis | R | R | S | 1 | >16 | ≤1 |
| C34 | 2014 | C | – | R | R | S | 1 | >16 | ≤1 |
| C35 | 2016 | N (SHH) | – | I | R | S | 0.5 | >16 | ≤1 |
| C36 | 2015 | N (SHH) | – | I | R | R | 0.5 | >16 | >32 |
| C37 | 2014 | N (SHH) | Albany | I | R | S | 0.5 | >16 | ≤1 |
| C38 | 2016 | N (SHH) | – | I | R | S | 0.25 | >16 | ≤1 |
| C39 | 2013 | N (SHH) | – | I | R | S | 0.25 | >16 | ≤1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fang, S.-B.; Lauderdale, T.-L.Y.; Huang, C.-H.; Chang, P.-R.; Wang, Y.-H.; Shigemura, K.; Lin, Y.-H.; Chang, W.-C.; Wang, K.-C.; Huang, T.-W.; et al. Genotypic Diversity of Ciprofloxacin Nonsusceptibility and Its Relationship with Minimum Inhibitory Concentrations in Nontyphoidal Salmonella Clinical Isolates in Taiwan. Antibiotics 2021, 10, 1383. https://doi.org/10.3390/antibiotics10111383
Fang S-B, Lauderdale T-LY, Huang C-H, Chang P-R, Wang Y-H, Shigemura K, Lin Y-H, Chang W-C, Wang K-C, Huang T-W, et al. Genotypic Diversity of Ciprofloxacin Nonsusceptibility and Its Relationship with Minimum Inhibitory Concentrations in Nontyphoidal Salmonella Clinical Isolates in Taiwan. Antibiotics. 2021; 10(11):1383. https://doi.org/10.3390/antibiotics10111383
Chicago/Turabian StyleFang, Shiuh-Bin, Tsai-Ling Yang Lauderdale, Chih-Hung Huang, Pei-Ru Chang, Yuan-Hung Wang, Katsumi Shigemura, Ying-Hsiu Lin, Wei-Chiao Chang, Ke-Chuan Wang, Tzu-Wen Huang, and et al. 2021. "Genotypic Diversity of Ciprofloxacin Nonsusceptibility and Its Relationship with Minimum Inhibitory Concentrations in Nontyphoidal Salmonella Clinical Isolates in Taiwan" Antibiotics 10, no. 11: 1383. https://doi.org/10.3390/antibiotics10111383
APA StyleFang, S. -B., Lauderdale, T. -L. Y., Huang, C. -H., Chang, P. -R., Wang, Y. -H., Shigemura, K., Lin, Y. -H., Chang, W. -C., Wang, K. -C., Huang, T. -W., & Chang, Y. -C. (2021). Genotypic Diversity of Ciprofloxacin Nonsusceptibility and Its Relationship with Minimum Inhibitory Concentrations in Nontyphoidal Salmonella Clinical Isolates in Taiwan. Antibiotics, 10(11), 1383. https://doi.org/10.3390/antibiotics10111383