Quantitative Evaluation of Nucleic Acid Degradability of Copper Alloy Surfaces and Its Correlation to Antibacterial Activity
Abstract
:1. Introduction
2. Results
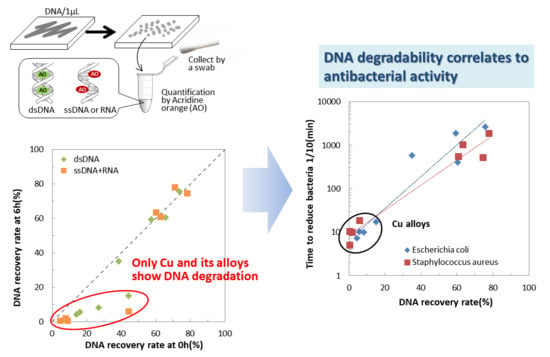
2.1. Nucleic Acid Degradability of Testing Materials
2.2. Nucleic Acid Degradation by Copper Salts
2.3. Antibacterial Activity of Testing Materials
2.4. Correlation between Nucleic Acid Degradability and Antibacterial Activity of Testing Materials
3. Discussion
3.1. Antibacterial Activity of Copper, Copper Alloys, and Other Antibacterial Materials
3.2. Nucleic Acid Degradability of Copper and Copper Alloys
4. Materials and Methods
4.1. A Testing Materials
4.2. Nucleic Acid Degradation Assay
4.3. Measurement of Copper Ion Release
4.4. Antibacterial Assay
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Review on Antimicrobial Resistance. Antimicrobial Resistance: Tackling a Crisis for the Health and Wealth of Nations; Review on Antimicrobial Resistance: London, UK, 2014. [Google Scholar]
- Weber, D.J.; Rutala, W.A. Understanding and Preventing Transmission of Healthcare-Associated Pathogens Due to the Contaminated Hospital Environment. Infect. Control Hosp. Epidemiol. 2013, 34, 449–452. [Google Scholar] [CrossRef] [Green Version]
- Kramer, A.; Schwebke, I.; Kampf, G. How long do nosocomial pathogens persist on inanimate surfaces? A systematic review. BMC. Infect. Dis. 2006, 6, 130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grass, G.; Rensing, C.; Solioz, M. Metallic copper as an antimicrobial surface. Appl. Environ. Microbiol. 2011, 77, 1541–1547. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borkow, G. Using Copper to Fight Microorganisms. Curr. Chem. Biol. 2012, 6, 93–103. [Google Scholar] [CrossRef]
- Michels, H.T.; Keevil, C.W.; Salgado, C.D.; Schmidt, M.G. From laboratory research to a clinical trial: Copper alloy surfaces kill bacteria and reduce hospital acquired infections. Health Environ. Res. Des. J. 2015, 9, 64–79. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, M.G.; Attaway, H.H.; Sharpe, P.A.; John, J.J.; Sepkowitz, K.A.; Morgan, A.; Fairey, S.E.; Singh, S.; Steed, L.L.; Cantey, J.R.; et al. Sustained reduction of microbial burden on common hospital surfaces through introduction of copper. J. Clin. Microbiol. 2012, 50, 2217–2223. [Google Scholar] [CrossRef] [Green Version]
- Salgado, C.D.; Sepkowitz, K.A.; John, J.F.; Cantey, J.R.; Attaway, H.H.; Freeman, K.D.; Sharpe, P.A.; Michels, H.T.; Schmidt, M.G. Copper surfaces reduce the rate of healthcare-acquired infections in the intensive care unit. Infect. Control Hosp. Epdemiol. 2013, 34, 479–486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pineda, I.; Hubbard, R.; Rodríguez, F. The role of copper surfaces in reducting the incidence of healthcare-associated infections: A systematic review and meta-analysis. Can. J. Infect. Control 2017, 32, 13–24. [Google Scholar]
- Schmidt, M.G.; von Dessauer, B.; Benavente, C.; Benadof, D.; Cifuentes, P.; Elgueta, A.; Duran, C.; Navarrete, M.S. Copper surfaces are associated with significantly lower concentrations of bacteria on selected surfaces within a paediatric intensive care unit. Am. J. Infect. Control 2016, 44, 203–209. [Google Scholar] [CrossRef] [Green Version]
- Michels, H.T.; Michels, C.A. Copper alloys—The new ‘old’ weapon in the fight against infectious disease. Curr. Trend. Microbiol. 2016, 10, 23–45. [Google Scholar]
- Weaver, L.; Noyce, J.O.; Michels, H.T.; Keevil, C.W. Potential action of copper surfaces on methicillin-resistant Staphylococcus aureus. J. Appl. Microbiol. 2010, 109, 2200–2205. [Google Scholar] [CrossRef] [PubMed]
- Santo, C.E.; Lam, E.W.; Elowsky, C.G.; Quaranta, D.; Domaille, D.W.; Chang, C.J.; Grass, G. Bacterial killing by dry metallic copper surfaces. Appl. Environ. Microbiol. 2011, 77, 794–802. [Google Scholar] [CrossRef] [Green Version]
- Hong, R.; Kang, T.Y.; Michels, C.A.; Gadura, N. Membrane lipid peroxidation in copper alloy-medicated contact killing of Escherichia coli. Appl. Environ. Microbiol. 2012, 78, 1776–1784. [Google Scholar] [CrossRef] [Green Version]
- Warnes, S.L.; Highmore, C.J.; Keevil, C.W. Horizontal transfer of antibiotic resistance genes on abiotic touch surfaces: Implications for public health. mBiol 2012, 3, e00489-12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Warnes, S.L.; Caves, V.; Keevil, C.W. Mechanism of copper surface toxicity in Escherichia coli O157:H7 and Salmonella involves immediate membrane depolarization followed by slower rate of DNA destruction which differs from that observed for Gram-positive bacteria. Environ. Microbiol. 2012, 14, 1730–1743. [Google Scholar] [CrossRef]
- Meyer, T.J.; Ramlall, J.; Thu, P.; Gadura, N. Antimicrobial properties of copper in gram-negative and gram-positive bacteria. Int. J. Pharmacol. Pharm. Sci. 2015, 9, 274–278. [Google Scholar]
- Warnes, S.L.; Keevil, C.W. Lack of involvement of Fenton chemistry in death of methicillin-resistant and methicillin-sensitive strains of Staphylococcus aureus and destruction of their genomes on wet or dry copper alloy surfaces. Appl. Environ. Microbiol. 2016, 82, 2132–2136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohkubo, N.; Nakamura, S.; Yamamoto, M.; Miyakusu, K.; Hasegawa, M. Antimaicrobial activity and basic properties of antimicrobial stainless steels “NSSAM series”. Nisshin Steel Tech. Rep. 1998, 77, 69–81. [Google Scholar]
- Hans, M.; Mathews, S.; Mücklich, F.; Solioz, M. Physicochemical properties of copper important for its antibacterial activity and development of a unified model. Biointerphases 2016, 11, 018902. [Google Scholar] [CrossRef]
- ISO 22196:2011. Measurement of Antibacterial Activity on Plastics and Other Non-Porous Surfaces; International Organization for Standardization: Geneva, Switzerland, 2011. [Google Scholar]
- Hiraishi, A.; Yoshida, N. An improved redox dye-staining method using 5-cyano-2, 3-ditoryl tetrazolium chloride for detection of metabolically active bacteria in activated sludge. Microbes Environ. 2004, 19, 61–70. [Google Scholar]
- Kitaguchi, A.; Yamaguchi, N.; Nasu, M. Enumeration of respiring pseudomonas spp. in milk within 6 hours by fluorescence in situ hybridization following formazan reduction. Appl. Environ. Microbiol. 2005, 71, 2748–2752. [Google Scholar] [CrossRef] [Green Version]
- Noyce, J.O.; Michels, H.; Keevil, C.W. Potential use of copper surfaces to reduce survival of epidemic methicillin-resistant Staphylococcus aureus in the healthcare environment. J. Hosp. Infect. 2006, 63, 289–297. [Google Scholar] [CrossRef] [PubMed]
- Noyce, J.O.; Michels, H.; Keevil, C.W. Use of copepr cast alloys to control Escherichia coli O157 cross-containation during food processing. Appl. Environ. Microbiol. 2006, 72, 4239–4244. [Google Scholar] [CrossRef] [Green Version]
- Japanese Industrial Standard JIS Z 2801:2012. Antimicrobial Products—Test for Antimicrobial Activity and Efficiency; Japan Standard Association: Tokyo, Japan, 2012. [Google Scholar]
- Matsunaga, T.; Tomoda, R.; Nakajima, T.; Wake, H. Photoelectrochemical sterilization of microbial cells by semiconductor powders. FEMS Microbiol. Lett. 1985, 29, 211–214. [Google Scholar] [CrossRef]
- Kikuchi, Y.; Sunada, K.; Iyoda, T.; Hashimoto, K.; Fujihima, A. Photocatalytic bactericidal effect of TiO2 thin films: Dynamic view of the active oxygen species responsible for the effect. J Photochem. Photobiol. A Chem. 1997, 106, 51–56. [Google Scholar] [CrossRef]
- Milošev, I.; Strehblow, H.H. Electrochemical behavior of Cu-xZn slloys in borate buffer solution at pH 9.2. J. Electrochem. Soc. 2003, 150, B517–B524. [Google Scholar] [CrossRef]
- Hans, M.; Erbe, A.; Mathews, S.; Chen, Y.; Solioz, M.; Mücklich, F. Role of copper oxides in contact killing of bacteria. Langmuir 2013, 29, 16160–16166. [Google Scholar] [CrossRef]
- Zeiger, M.; Solioz, M.; Edongue, H.; Arzt, E.; Schneider, A.S. Surface structure influences contact killing of bacteria by copper. Mcrobiol. Open 2014, 3, 327–332. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, S.; Miyakusu, K.; Sato, Y.; Kikuchi, Y.; Kawakami, H. Antimicrobiability of Cu contained stainless steels. Tetsu-Hagané 2014, 100, 1021–1028. [Google Scholar] [CrossRef] [Green Version]
- Notoya, T. Corrosion and corrosion resistance of copper. Pip. Eng. 2004, 46, 1–7. [Google Scholar]
- Fujiwara, S. Why do nucleic acids denature? Seibutsu-Kogaku Kaishi 2011, 89, 200–203. [Google Scholar]
- Sigel, R.K.O.; Sigel, H. A stability concept for metal ion coordination to single-stranded nucleic acids and affinities of individual sites. Acc. Chem. Res. 2010, 43, 974–984. [Google Scholar] [CrossRef] [Green Version]
- Warens, S.L.; Keevil, C.W. Inactivation of Norovirus on dry copper alloy surfaces. PLoS ONE 2013, 8, e75017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Warens, S.L.; Little, Z.R.; Keevil, C.W. Human Coronavirus 229E remains infectious on common touch surface materials. mBiol 2015, 6, e01697-15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manuel, C.S.; Moore, M.D.; Jaykus, L.A. Destruction of the capsid and genome of GII.4 human Norovirus occurs during exposure to metal alloys containing copper. Appl. Environ. Microbiol. 2015, 81, 4940–4946. [Google Scholar] [CrossRef] [PubMed] [Green Version]










| C1020 | C6932 | CBRA | CBRI | ABSS | Ag | Glass | Resin X | Resin Y | ||
|---|---|---|---|---|---|---|---|---|---|---|
| E.coli | T1/10 | 6.27 | 9.97 | 10.2 | 15.7 | 383 | 575 | 1230 | 1830 | 2670 |
| T1/100 | 9.33 | 24.6 | 23.4 | 34.6 | 1490 | 1990 | 4060 | 10,500 | 20,400 | |
| S.aureus | T1/10 | 5.30 | 10.3 | 9.89 | 15.8 | 504 | 530 | 1120 | 1000 | 1850 |
| T1/100 | 9.86 | 25.0 | 23.8 | 30.7 | 2030 | 1780 | 5280 | 4630 | 10,900 |
| Sample | Cu | Zn | Sn | Ni | Mn | Cr | Fe | Si | P |
|---|---|---|---|---|---|---|---|---|---|
| C1020 | >99.99 | ||||||||
| C6932 | 75.4 | Rem. | 3.1 | 0.09 | |||||
| CBRA | 70.1 | Rem. | 0.5 | 2.0 | |||||
| CBRI | 54.0 | Rem. | 0.4 | 10.9 | |||||
| ABSS | 3.8 | 9.4 | 1.4 | 18.1 | Rem. | 0.6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yamamoto, A.; Tanaka, S.; Ohishi, K. Quantitative Evaluation of Nucleic Acid Degradability of Copper Alloy Surfaces and Its Correlation to Antibacterial Activity. Antibiotics 2021, 10, 1439. https://doi.org/10.3390/antibiotics10121439
Yamamoto A, Tanaka S, Ohishi K. Quantitative Evaluation of Nucleic Acid Degradability of Copper Alloy Surfaces and Its Correlation to Antibacterial Activity. Antibiotics. 2021; 10(12):1439. https://doi.org/10.3390/antibiotics10121439
Chicago/Turabian StyleYamamoto, Akiko, Shinji Tanaka, and Keiichiro Ohishi. 2021. "Quantitative Evaluation of Nucleic Acid Degradability of Copper Alloy Surfaces and Its Correlation to Antibacterial Activity" Antibiotics 10, no. 12: 1439. https://doi.org/10.3390/antibiotics10121439
APA StyleYamamoto, A., Tanaka, S., & Ohishi, K. (2021). Quantitative Evaluation of Nucleic Acid Degradability of Copper Alloy Surfaces and Its Correlation to Antibacterial Activity. Antibiotics, 10(12), 1439. https://doi.org/10.3390/antibiotics10121439