Dual Roles of Extracellular Histone H3 in Host Defense: Its Differential Regions Responsible for Antimicrobial and Cytotoxic Properties and Their Modes of Action
Abstract
:1. Introduction
2. Experimental Results
2.1. Antimicrobial Activities of the Full-Length Histone H3 and SEM Analysis
2.2. Cytotoxic Activities of the Full-Length Histone H3 and SEM Analysis
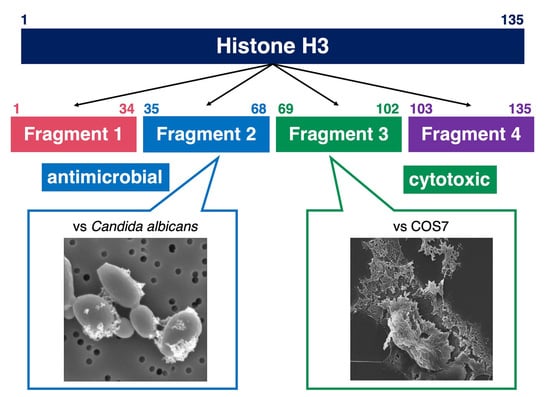
2.3. Identification of Antimicrobial and Cytotoxic Regions of Histone H3
2.4. Bacterial Endotoxin Binding Ability of Histone H3 and Its Related Peptides
3. Discussion
4. Materials and Methods
4.1. Synthetic Peptides
4.2. Bacterial and Fungal Cell Strains
4.3. Mammalian Cell Lines
4.4. Antimicrobial Assay
4.5. Cytotoxic Assay
4.6. Scanning Electron Microscopy
4.7. Enzyme-Linked Endotoxin Binding Assay
4.8. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Campos, E.; Reinberg, D. Histones: Annotating chromatin. Annu. Rev. Genet. 2009, 43, 559–599. [Google Scholar] [CrossRef] [PubMed]
- Zentner, G.E.; Henikoff, S. Regulation of nucleosome dynamics by histone modifications. Nat. Struct. Mol. Biol. 2013, 20, 259–266. [Google Scholar] [CrossRef] [PubMed]
- Dráb, T.; Kračmerová, J.; Hanzlíková, E.; Černá, T.; Litváková, R.; Pohlová, A.; Tichá, M.; Přikryl, P.; Liberda, J. The antimicrobial action of histones in the reproductive tract of cow. Biochem. Biophys. Res. Commun. 2014, 443, 987–990. [Google Scholar] [CrossRef]
- Hoeksema, M.; van Eijk, M.; Haagsman, H.P.; Hartshorn, K.L. Histones as mediators of host defense, inflammation and thrombosis. Future Microbiol. 2016, 11, 441–453. [Google Scholar] [CrossRef] [PubMed]
- Miller, B.F.; Abrams, R.; Dorfman, A.; Klein, M. Antibacterial properties of protamine and histone. Science 1942, 96, 428–430. [Google Scholar] [CrossRef]
- Patat, S.A.; Carnegie, R.B.; Kingsbury, C.; Gross, P.S.; Chapman, R.; Schey, K.L. Antimicrobial activity of histones from hemocytes of the Pacific white shrimp. Eur. J. Biochem. 2004, 271, 4825–4833. [Google Scholar] [CrossRef]
- Park, I.Y.; Park, C.B.; Kim, M.S.; Kim, S.C. Parasin I, an antimicrobial peptide derived from histone H2A in the cat fish, Parasilurus asotus. FEBS Lett. 1998, 437, 258–262. [Google Scholar] [CrossRef]
- Richards, R.C.; O’Neil, D.B.; Thibault, P.; Ewart, K.V. Histone H1: An antimicrobial protein of Atlantic salmon (Salmo salar). Biochem. Biophys. Res. Commun. 2001, 284, 549–555. [Google Scholar] [CrossRef]
- Noga, E.J.; Fan, Z.; Silphaduang, U. Histone-like proteins from fish are lethal to the parasitic dinoflagellate Amyloodinium ocellatum. Parasitology 2001, 123, 57–65. [Google Scholar] [CrossRef]
- Nam, B.H.; Seo, J.K.; Go, H.J.; Lee, M.J.; Kim, Y.O.; Kim, D.G.; Lee, S.J.; Park, N.G. Purification and characterization of an antimicrobial histone H1-like protein and its gene from the testes of olive flounder, Paralichthys olivaceus. Fish Shellfish Immunol. 2012, 33, 92–98. [Google Scholar] [CrossRef]
- Park, C.B.; Kim, M.S.; Kim, S.C. A novel antimicrobial peptide from Bufo bufo gargarizans. Biochem. Biophys. Res. Commun. 1996, 218, 408–413. [Google Scholar] [CrossRef] [PubMed]
- Kawasaki, H.; Koyama, T.; Conlon, J.M.; Yamakura, F.; Iwamuro, S. Antimicrobial action of histone H2B in Escherichia coli: Evidence for membrane translocation and DNA-binding of a histone H2B fragment after proteolytic cleavage by outer membrane proteinase T. Biochimie 2008, 90, 1693–1702. [Google Scholar] [CrossRef] [PubMed]
- Silphaduang, U.; Hincke, M.T.; Nys, Y.; Mine, Y. Antimicrobial proteins in chicken reproductive system. Biochem. Biophys. Res. Commun. 2005, 340, 648–655. [Google Scholar] [CrossRef] [PubMed]
- Li, G.H.; Mine, Y.; Hincke, M.T.; Nys, Y. Isolation and characterization of antimicrobial proteins and peptide from chicken liver. J. Pept. Sci. 2007, 13, 368–378. [Google Scholar] [CrossRef]
- Rose, F.R.; Bailey, K.; Keyte, J.W.; Chan, W.C.; Greenwood, D.; Mahida, Y.R. Potential role of epithelial cell-derived histone H1 proteins in innate antimicrobial defense in the human gastrointestinal tract. Infect. Immun. 1998, 66, 3255–3263. [Google Scholar] [CrossRef]
- Kim, H.S.; Cho, J.H.; Park, H.W.; Yoon, H.; Kim, M.S.; Kim, S.C. Endotoxin-neutralizing antimicrobial proteins of the human placenta. J. Immunol. 2002, 168, 2356–2364. [Google Scholar] [CrossRef]
- Lee, D.Y.; Huang, C.M.; Nakatsuji, T.; Thiboutot, D.; Kang, S.A.; Monestier, M.; Gallo, R.L. Histone H4 is a major component of the antimicrobial action of human sebocytes. J. Investig. Dermatol. 2009, 129, 2489–2496. [Google Scholar] [CrossRef]
- De Lucca, A.J.; Heden, L.O.; Ingber, B.; Bhatnagar, D.J. Antifungal Properties of Wheat Histones (H1–H4) and Purified Wheat Histone H1. Agric. Food Chem. 2011, 59, 6933–6939. [Google Scholar] [CrossRef]
- Kawasaki, H.; Iwamuro, S. Potential roles of histones in host defense as antimicrobial agents. Infect. Disord. Drug Targets 2008, 8, 195–205. [Google Scholar] [CrossRef]
- Li, Y.; Wan, D.; Luo, X.; Song, T.; Wang, Y.; Yu, Q.; Jiang, L.; Liao, R.; Zhao, W.; Su, B. Circulating histones in sepsis: Potential outcome predictors and therapeutic targets. Front. Immunol. 2021, 12, 650184. [Google Scholar] [CrossRef]
- Tagai, C.; Morita, S.; Shiraishi, T.; Miyaji, K.; Iwamuro, S. Antimicrobial properties of arginine- and lysine-rich histones and involvement of bacterial outer membrane protease T in their differential mode of actions. Peptides 2011, 32, 2003–2009. [Google Scholar] [CrossRef] [PubMed]
- Morita, S.; Tagai, C.; Shiraishi, T.; Miyaji, K.; Iwamuro, S. Differential mode of antimicrobial actions of arginine-rich and lysine-rich histones against Gram-positive Staphylococcus aureus. Peptides 2013, 48, 75–82. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.S.; Yoon, H.; Minn, I.; Park, C.B.; Lee, W.T.; Zasloff, M.; Kim, S.C. Pepsin-mediated processing of the cytoplasmic histone H2A to strong antimicrobial peptide buforin I. J. Immunol. 2000, 165, 3268–3274. [Google Scholar] [CrossRef] [PubMed]
- Tsao, H.S.; Spinella, S.A.; Lee, A.T.; Elmore, D.E. Design of novel histone-derived antimicrobial peptides. Peptides 2009, 30, 2168–2173. [Google Scholar] [CrossRef]
- Pavia, K.E.; Spinella, S.A.; Elmore, D.E. Novel histone-derived antimicrobial peptides use different antimicrobial mechanisms. Biochim. Biophys. Acta 2012, 1818, 869–876. [Google Scholar] [CrossRef] [PubMed]
- Jodoin, J.; Hincke, M.T. Histone H5 is a potent antimicrobial agent and a template for novel antimicrobial peptides. Sci. Rep. 2018, 8, 2411. [Google Scholar] [CrossRef]
- Brinkmann, V.; Reichard, U.; Goosmann, C.; Fauler, B.; Uhlemann, Y.; Weiss, D.S.; Weinrauch, Y.; Zychlinsky, A. Neutrophil extracellular traps kill bacteria. Science 2004, 303, 1532–1535. [Google Scholar] [CrossRef]
- Fuchs, T.A.; Brill, A.; Duerschmied, D.; Schatzberg, D.; Monestier, M.; Myers, D.D., Jr.; Wrobleski, S.K.; Wakefield, T.W.; Hartwig, J.H.; Wagner, D.D. Extracellular DNA traps promote thrombosis. Proc. Natl. Acad. Sci. USA 2010, 107, 15880–15885. [Google Scholar] [CrossRef]
- Urban, C.F.; Ermert, D.; Schmid, M.; Abu-Abed, U.; Goosmann, C.; Nacken, W.; Brinkmann, V.; Jungblut, P.R.; Zychlinsky, A. Neutrophil extracellular traps contain calprotectin, a cytosolic protein complex involved in host defense against Candida albicans. PLoS Pathog. 2009, 5, e1000639. [Google Scholar] [CrossRef]
- Xu, J.; Zhang, X.; Pelayo, R.; Monestier, M.; Ammollo, C.T.; Semeraro, F.; Taylor, F.B.; Esmon, N.L.; Lupu, F.; Esmon, C.T. Extracellular histones are major mediators of death in sepsis. Nat. Med. 2009, 15, 1318–1321. [Google Scholar] [CrossRef] [Green Version]
- Abrams, S.T.; Zhang, N.; Dart, C.; Wang, S.S.; Thachil, J.; Guan, Y.; Wang, G.; Toh, C.H. Human CRP defends against the toxicity of circulating histones. J. Immunol. 2013, 191, 2495–2502. [Google Scholar] [CrossRef] [PubMed]
- Abrams, S.T.; Zhang, N.; Manson, J.; Liu, T.; Dart, C.; Baluwa, F.; Wang, S.S.; Brohi, K.; Kipar, A.; Yu, W.; et al. Circulating histones are mediators of trauma-associated lung injury. Am. J. Respir. Crit. Care Med. 2013, 187, 160–169. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Zhang, X.; Monestier, M.; Esmon, N.L.; Esmon, C.T. Extracellular histones are mediators of death through TLR2 and TLR4 in mouse fatal liver injury. J. Immunol. 2011, 187, 2626–2631. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Li, L.; Liu, J.; Lv, B.; Chen, F. Extracellular histones induce tissue factor expression in vascular endothelial cells via TLR and activation of NF-κB and AP-1. Thromb. Res. 2016, 137, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Silk, E.; Zhao, H.; Weng, H.; Ma, D. The role of extracellular histone in organ injury. Cell Death Dis. 2018, 8, e2812. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, Y.; Ito, T.; Yasuda, T.; Furubeppu, H.; Kamikokuryo, C.; Yamada, S.; Maruyama, I.; Kakihana, Y. Circulating histone H3 levels in septic patients are associated with coagulopathy, multiple organ failure, and death: A single-center observational study. Thromb. J. 2019, 17, 1. [Google Scholar] [CrossRef]
- Martín-Rodríguez, A.J.; González-Orive, A.; Hernández-Creus, A.; Morales, A.; Dorta-Guerra, R.; Norte, M.; Martín, V.S.; Fernández, J.J. On the influence of the culture conditions in bacterial antifouling bioassays and biofilm properties: Shewanella algae, a case study. BMC Microbiol. 2014, 14, 102. [Google Scholar] [CrossRef]
- Gibson, B.; Wilson, D.J.; Feil, E.; Eyre-Walker, A. The distribution of bacterial doubling times in the wild. Proc. Biol. Sci. 2018, 285, 20180789. [Google Scholar] [CrossRef]
- Zasloff, M. Antimicrobial peptides of multicellular organisms. Nature 2002, 415, 389–395. [Google Scholar] [CrossRef]
- Brogden, K.A.; Ackermann, M.; McCray, P.B., Jr.; Tack, B.F. Antimicrobial peptides in animals and their role in host defences. Int. J. Antimicrob. Agents 2003, 22, 465–478. [Google Scholar] [CrossRef] [Green Version]
- Brogden, K.A. Antimicrobial peptides: Pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 2005, 3, 238–250. [Google Scholar] [CrossRef] [PubMed]
- Iba, T.; Hashiguchi, N.; Nagaoka, I.; Tabe, Y.; Kadota, K.; Sato, K. Heparins attenuated histone-mediated cytotoxicity in vitro and improved the survival in a rat model of histone-induced organ dysfunction. Intensive. Care Med. Exp. 2015, 3, 36. [Google Scholar] [CrossRef] [PubMed]
- Kutcher, M.E.; Xu, J.; Vilardi, R.F.; Ho, C.; Esmon, C.T.; Cohen, M.J. Extracellular histone release in response to traumatic injury: Implications for a compensatory role of activated protein C. J. Trauma Acute Care Surg. 2012, 73, 1389–1394. [Google Scholar] [CrossRef] [PubMed]
- Marsman, G.; von Richthofen, H.; Bulder, I.; Lupu, F.; Hazelzet, J.; Luken, B.M.; Zeerleder, S. DNA and factor VII-activating protease protect against the cytotoxicity of histones. Blood Adv. 2017, 1, 2491–2502. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, L.; Cao, C.; Jin, H.; Zhang, Y.; Liu, Y.; Gao, Y.; Liang, X.; Li, G.; Shou, S. Heparin attenuates histone-mediated cytotoxicity in septic acute kidney injury. Front. Med. 2020, 7, 586652. [Google Scholar] [CrossRef]
- Ogawa, D.; Suzuki, M.; Inamura, Y.; Saito, K.; Hasunuma, I.; Kobayashi, T.; Kikuyama, S.; Iwamuro, S. Antimicrobial property and mode of action of the skin peptides of the Sado wrinkled frog, Glandirana susurra, against animal and plant pathogens. Antibiotics 2020, 9, 457. [Google Scholar] [CrossRef]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tanaka, Y.; Yamanaka, N.; Koyano, I.; Hasunuma, I.; Kobayashi, T.; Kikuyama, S.; Iwamuro, S. Dual Roles of Extracellular Histone H3 in Host Defense: Its Differential Regions Responsible for Antimicrobial and Cytotoxic Properties and Their Modes of Action. Antibiotics 2022, 11, 1240. https://doi.org/10.3390/antibiotics11091240
Tanaka Y, Yamanaka N, Koyano I, Hasunuma I, Kobayashi T, Kikuyama S, Iwamuro S. Dual Roles of Extracellular Histone H3 in Host Defense: Its Differential Regions Responsible for Antimicrobial and Cytotoxic Properties and Their Modes of Action. Antibiotics. 2022; 11(9):1240. https://doi.org/10.3390/antibiotics11091240
Chicago/Turabian StyleTanaka, Yuri, Nanako Yamanaka, Izumi Koyano, Itaru Hasunuma, Tetsuya Kobayashi, Sakae Kikuyama, and Shawichi Iwamuro. 2022. "Dual Roles of Extracellular Histone H3 in Host Defense: Its Differential Regions Responsible for Antimicrobial and Cytotoxic Properties and Their Modes of Action" Antibiotics 11, no. 9: 1240. https://doi.org/10.3390/antibiotics11091240
APA StyleTanaka, Y., Yamanaka, N., Koyano, I., Hasunuma, I., Kobayashi, T., Kikuyama, S., & Iwamuro, S. (2022). Dual Roles of Extracellular Histone H3 in Host Defense: Its Differential Regions Responsible for Antimicrobial and Cytotoxic Properties and Their Modes of Action. Antibiotics, 11(9), 1240. https://doi.org/10.3390/antibiotics11091240