Structure–Activity Relationship Studies of Substitutions of Cationic Amino Acid Residues on Antimicrobial Peptides
Abstract
:1. Introduction
2. Results
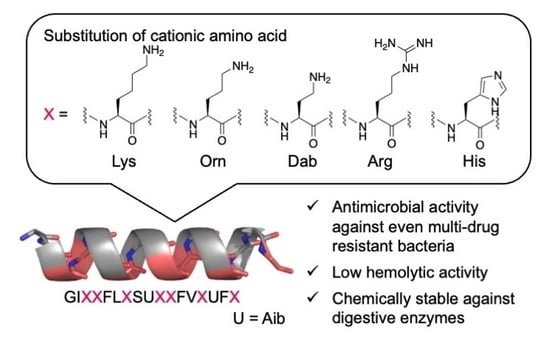
2.1. Design and Synthesis of AMPs
2.2. Preferred Secondary Structures of the Synthesized Peptides
2.3. Antimicrobial Activity and Hemolytic Activity
2.4. Chemical Stability of the Peptides against Digestive Enzymes
3. Discussion
4. Materials and Methods
4.1. General Information
4.2. Peptide Synthesis
4.3. CD Spectrometry
4.4. Antimicrobial Activity
4.5. Digestion Assay
4.6. Hemolysis Activity
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hutchings, M.I.; Truman, A.W.; Wilkinon, B. Antibiotics: Past, present and future. Curr. Opin. Microbiol. 2019, 51, 72–80. [Google Scholar] [CrossRef] [PubMed]
- Blair, J.M.A.; Webber, M.A.; Baylay, A.J.; Ogbolu, D.O.; Piddock, L.J.V. Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 2015, 13, 42–51. [Google Scholar] [CrossRef] [PubMed]
- Da Cunha, N.B.; Cobacho, N.B.; Viana, J.F.C.; Lima, L.A.; Sampaio, K.B.O.; Dohms, S.S.M.; Ferreira, A.C.R.; de la Fuente-Núñez, C.; Costa, F.F.; Franco, O.L.; et al. The next generation of antimicrobial peptides (AMPs) as molecular therapeutic tools for the treatment of diseases with social and economic impacts. Drug Discov. Today 2017, 22, 234–248. [Google Scholar] [CrossRef] [PubMed]
- Ageitos, J.M.; Sánchez-Pérez, A.; Calo-Meta, P.; Villa, T.G. Antimicrobial peptides (amps): Ancient compounds that represent novel weapons in the fight against bacteria. Biochem. Pharmacol. 2017, 133, 117–138. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Zhang, X.; Yuan, Y.; Bao, Y.; Xiong, M. Role and modulation of the secondary structure of antimicrobial peptides to improve selectivity. Biomater. Sci. 2020, 8, 6858–6866. [Google Scholar] [CrossRef]
- Elliott, A.G.; Huang, J.X.; Neve, S.; Zuegg, J.; Edwards, I.A.; Cain, A.K.; Boinett, C.J.; Barquist, L.; Lundberg, C.V.; Steen, J.; et al. An amphipathic peptide with antibiotic activity against multidrug-resistant Gram-negative bacteria. Nat. Commun. 2020, 11, 3184. [Google Scholar] [CrossRef]
- Yokoo, H.; Hirano, M.; Misawa, T.; Demizu, Y. Helical antimicrobial peptide foldamers containing non-proteinogenic amino acids. ChemMedChem 2021, 16, 1226–1233. [Google Scholar] [CrossRef]
- Tew, G.N.; Scott, R.W.; Klein, M.L.; DeGrado, W.F. De Novo design of antimicrobial polymers, foldamers, and small molecules: From discovery to practical applications. Acc. Chem. Res. 2011, 43, 30–39. [Google Scholar] [CrossRef] [Green Version]
- Bonnel, C.; Legrand, B.; Simon, M.; Clavié, M.; Masnou, A.; Jumas-Bilak, E.; Kang, Y.K.; Licznar-Fajardo, P.; Maillard, L.T.; Masurier, N. Tailoring the physicochemical properties of antimicrobial peptides onto a thiazole-based γ-peptide foldamer. J. Med. Chem. 2020, 63, 9168–9180. [Google Scholar] [CrossRef]
- Horne, W.S.; Gellman, S.H. Foldamers with heterogenous backbones. Acc. Chem. Res. 2008, 41, 1299–1408. [Google Scholar] [CrossRef]
- Porter, E.A.; Wang, X.; Lee, H.S.; Weisblum, B.; Gellman, S.H. Non-haemolytic beta-amino-acid oligomers. Nature 2000, 404, 565. [Google Scholar] [CrossRef] [PubMed]
- Mourtada, R.; Herce, H.D.; Yin, D.J.; Moroco, J.A.; Wales, T.E.; Engen, J.R.; Walensky, L.D. Design of stapled antimicrobial peptides that are stable, nontoxic and kill antibiotic-resistant bacteria in mice. Nat. Biotechnol. 2019, 37, 1186–1197. [Google Scholar] [CrossRef] [PubMed]
- Goto, C.; Hirano, M.; Hayashi, K.; Kikuchi, Y.; Hara-Kudo, Y.; Misawa, T.; Demizu, Y. Development of amphipathic antimicrobial peptide foldamers based on Magainin 2 sequence. ChemMedChem 2019, 14, 1911–1916. [Google Scholar] [CrossRef] [PubMed]
- Hirano, M.; Saito, C.; Yokoo, H.; Goto, C.; Kawano, R.; Misawa, T.; Demizu, Y. Development of antimicrobial stapled peptides based on Magainin 2 sequence. Molecules 2021, 26, 444. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.K.; Kim, D.G.; Lee, J.E.; Park, K.S.; Lee, I.A.; Lee, K.Y.; Kim, Y.O.; Nam, B.H. Antimicrobial activity and action mechanisms of Argrich short analog peptides designed from the C-terminal loop region of American oyster defensin (AOD). Mar. Drugs 2021, 19, 451. [Google Scholar] [CrossRef] [PubMed]
- Dong, N.; Wang, C.; Zhang, T.; Zhang, L.; Xue, C.; Feng, X.; Bi, C.; Shan, A. Bioactivity and bactericidal mechanism of Histidine-rich β-hairpin peptide against Gram-negative bacteria. Int. J. Mol. Sci. 2019, 20, 3954. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frenkel-Pinter, M.; Haynes, J.W.; Martin, C.; Petrov, A.S.; Burcar, B.T.; Krishnamurthy, R.; Hudm, N.V.; Leman, L.J.; Williams, L.D. Selective incorporation of proteinaceous over nonproteinaceous cationic amino acids in model prebiotic oligomerization reactions. Proc. Natl. Acad. Sci. USA 2019, 116, 16338–16346. [Google Scholar] [CrossRef] [Green Version]
- Kelly, S.M.; Price, N.C. The use of circular dichroism in the investigation of protein structure and function. Curr. Protein Pept. Sci. 2000, 1, 349–384. [Google Scholar] [CrossRef] [Green Version]
- Su, Y.; Doherty, T.; Waring, A.J.; Ruchala, P.; Hong, M. Roles of arginine and lysine residues in the translocation of a cell-penetrating peptide from (13)C, (31)P, and (19)F solid-state NMR. Biochemistry 2009, 48, 4587–4595. [Google Scholar] [CrossRef] [Green Version]
- Mergler, M.; Dick, F.; Sax, B.; Schwindling, J.; Vorherr, T.H. Synthesis and application of Fmoc-His(3-Bum)-OH. J. Pept. Sci. 2001, 7, 502–510. [Google Scholar] [CrossRef]
- Sandín, D.; Valle, J.; Chaves-Arquero, B.; Prats-Ejarque, G.; Larrosa, M.N.; González-López, J.J.; Jiménez, M.Á.; Boix, E.; Andreu, D.; Torrent, M. Rationally modified antimicrobial peptides from the N-terminal domain of human RNase 3 show exceptional serum stability. J. Med. Chem. 2021, 64, 11472–11482. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Guarnieri, M.T.; Vasil, A.I.; Vasil, M.L.; Mant, C.T.; Hodges, R.S. Role of peptide hydrophobicity in the mechanism of action of alpha-helical antimicrobial peptides. Antimicrob. Agents Chemother. 2007, 51, 1398–1406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horne, W.S.; Boersma, M.D.; Windsor, M.A.; Gellman, S.H. Sequence-based design of alpha/beta-peptide foldamers that mimic BH3 domains. Angew. Chem. Int. Ed. Engl. 2008, 47, 2853–2856. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pasco, M.; Dolain, C.; Guichard, G. Foldamers in Medicinal Chemistry. Compr. Supramol. Chem. II 2017, 89–125. [Google Scholar]
- Cavaco, M.; Andreu, D.; Castanho, M.A.R.B. The challenge of peptide proteolytic stability studies: Scarce data, difficult readability, and the need for harmonization. Angew. Chem. Int. Ed. Engl. 2021, 60, 1686–1688. [Google Scholar] [CrossRef]
- Akishiba, M.; Takeuchi, T.; Kawaguchi, Y.; Sakamoto, K.; Yu, H.H.; Nakase, I.; Takatani-Nakase, T.; Madani, F.; Gräslund, A.; Futaki, S. Cytosolic antibody delivery by lipid-sensitive endosomolytic peptide. Nat. Chem. 2017, 9, 751–761. [Google Scholar] [CrossRef]



| Peptide | Sequence | |
|---|---|---|
| Mag2 | H-GIGKFLHSAKKFGKAFVGEIMNS-NH2 |  |
| 1 | H-GIKKFLKSXKKFVKXFK-NH2 | |
| 2 | H-GIOOFLOSUOOFVOUFO-NH2 | |
| 3 | H-GIBBFLBSUBBFVBUFB-NH2 | |
| 4 | H-GIRRFLRSURRFVRUFR-NH2 | |
| 5 | H-GIHHFLHSUHHFVHUFH-NH2 |
| Peptide | Sequence | MIC (µM) | Hemolysis (µM) | |||
|---|---|---|---|---|---|---|
| Gram (–) | Gram (+) | |||||
| E.Coli | P.a. | MDRP | S.a. | |||
| Mag2 | H-GIGKFLHSAKKFGKAFVGEIMNS-NH2 | 3.13 | 12.5 | 12.5 | 100 | >100 |
| 1 | H-GIKKFLKSXKKFVKXFK-NH2 | 3.13 | 3.13 | 3.13 | 12.5 | 100 |
| 2 | H-GIOOFLOSUOOFVOUFO-NH2 | 3.13 | 6.25 | 3.13 | 12.5 | 50 |
| 3 | H-GIBBFLBSUBBFVBUFB-NH2 | 3.13 | 6.25 | 6.25 | 12.5 | 25 |
| 4 | H-GIRRFLRSURRFVRUFR-NH2 | 3.13 | 12.5 | 12.5 | 3.13 | 6.25 |
| 5 | H-GIHHFLHSUHHFVHUFH-NH2 | >50 | >50 | >50 | >50 | >100 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Takada, M.; Ito, T.; Kurashima, M.; Matsunaga, N.; Demizu, Y.; Misawa, T. Structure–Activity Relationship Studies of Substitutions of Cationic Amino Acid Residues on Antimicrobial Peptides. Antibiotics 2023, 12, 19. https://doi.org/10.3390/antibiotics12010019
Takada M, Ito T, Kurashima M, Matsunaga N, Demizu Y, Misawa T. Structure–Activity Relationship Studies of Substitutions of Cationic Amino Acid Residues on Antimicrobial Peptides. Antibiotics. 2023; 12(1):19. https://doi.org/10.3390/antibiotics12010019
Chicago/Turabian StyleTakada, Mayu, Takahito Ito, Megumi Kurashima, Natsumi Matsunaga, Yosuke Demizu, and Takashi Misawa. 2023. "Structure–Activity Relationship Studies of Substitutions of Cationic Amino Acid Residues on Antimicrobial Peptides" Antibiotics 12, no. 1: 19. https://doi.org/10.3390/antibiotics12010019
APA StyleTakada, M., Ito, T., Kurashima, M., Matsunaga, N., Demizu, Y., & Misawa, T. (2023). Structure–Activity Relationship Studies of Substitutions of Cationic Amino Acid Residues on Antimicrobial Peptides. Antibiotics, 12(1), 19. https://doi.org/10.3390/antibiotics12010019