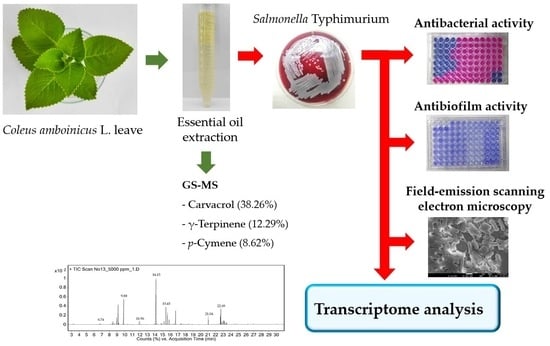
Transcriptional Profiling of the Effect of Coleus amboinicus L. Essential Oil against Salmonella Typhimurium Biofilm Formation
Abstract
:1. Introduction
2. Results
2.1. Extraction and Composition of the Essential Oil
2.2. Determination of Antimicrobial and Antibiofilm Activities of EO-CA
2.3. Global Changes at the Transcriptome Level
2.4. Analysis of DEGs
2.5. qRT-PCR Validation of Gene Expression
2.6. Field-Emission Scanning Electron Microscopy
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. Bacterial Strain and Culture
4.3. Antimicrobial Activity of EO-CA
4.4. Antibiofilm Formation of EO-CA
4.5. Crystal Violet Staining
4.6. FESEM
4.7. RNA Extraction
4.8. Transcriptomics Analysis
4.9. Gene Expression Analysis
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- CDC. Salmonella Outbreak Linked to Onions. Available online: https://www.cdc.gov/salmonella/index.html (accessed on 10 August 2023).
- Harrell, J.E.; Hahn, M.M.; D’Souza, S.J.; Vasicek, E.M.; Sandala, J.L.; Gunn, J.S.; McLachlan, J.B. Salmonella biofilm formation, chronic infection, and immunity within the intestine and hepatobiliary tract. Front. Cell Infect. Microbiol. 2020, 10, 624622. [Google Scholar] [CrossRef] [PubMed]
- Knodler, L.A.; Elfenbein, J.R. Salmonella enterica. Trends Microbiol. 2019, 27, 964–965. [Google Scholar] [CrossRef] [PubMed]
- Fàbrega, A.; Vila, J. Salmonella enterica serovar Typhimurium skills to succeed in the host: Virulence and regulation. Clin. Microbiol. Rev. 2013, 26, 308–341. [Google Scholar] [CrossRef] [PubMed]
- Gildea, L.; Ayariga, J.A.; Xu, J.; Villafane, R.; Robertson, B.K.; Samuel-Foo, M.; Ajayi, O.S. Cannabis sativa CBD extract exhibits synergy with broad-spectrum antibiotics against Salmonella enterica subsp. Enterica serovar Typhimurium. Microorganisms 2022, 10, 2360. [Google Scholar] [CrossRef]
- Gildea, L.; Ayariga, J.A.; Ajayi, O.S.; Xu, J.; Villafane, R.; Samuel-Foo, M. Cannabis sativa CBD extract shows promising antibacterial activity against Salmonella Typhimurium and S. newington. Molecules 2022, 27, 2669. [Google Scholar] [CrossRef]
- Ferrari, R.G.; Rosario, D.K.A.; Cunha-Neto, A.; Mano, S.B.; Figueiredo, E.E.S.; Conte-Junior, C.A. Worldwide epidemiology of Salmonella serovars in animal-based foods: A meta-analysis. Appl. Environ. Microbiol. 2019, 85, e00591-19. [Google Scholar] [CrossRef]
- Nazari Moghadam, M.M.; Rahimi, E.; Shakerian, A.; Momtaz, H. Prevalence of Salmonella Typhimurium and Salmonella Enteritidis isolated from poultry meat: Virulence and antimicrobial-resistant genes. BMC Microbiol. 2023, 23, 168. [Google Scholar] [CrossRef]
- Ehuwa, O.; Jaiswal, A.K.; Jaiswal, S. Salmonella, food safety and food handling practices. Foods 2021, 10, 907. [Google Scholar] [CrossRef]
- Liu, X.; Yao, H.; Zhao, X.; Ge, C. Biofilm formation and control of foodborne pathogenic bacteria. Molecules 2023, 28, 2432. [Google Scholar] [CrossRef]
- Steenackers, H.; Hermans, K.; Vanderleyden, J.; De Keersmaecker, S.C.J. Salmonella biofilms: An overview on occurrence, structure, regulation and eradication. Food Res. Int. 2012, 45, 502–531. [Google Scholar] [CrossRef]
- Karygianni, L.; Ren, Z.; Koo, H.; Thurnheer, T. Biofilm matrixome: Extracellular components in structured microbial communities. Trends Microbiol. 2020, 28, 668–681. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Liu, B.; Ding, X.; Bin, P.; Yang, Y.; Zhu, G. Regulatory mechanisms between quorum sensing and virulence in Salmonella. Microorganisms 2022, 10, 2211. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Jyung, S.; Kang, D.H. Comparative study of Salmonella Typhimurium biofilms and their resistance depending on cellulose secretion and maturation temperatures. LWT 2022, 154, 112700. [Google Scholar] [CrossRef]
- Galié, S.; García-Gutiérrez, C.; Miguélez, E.M.; Villar, C.J.; Lombó, F. Biofilms in the food industry: Health aspects and control methods. Front. Microbiol. 2018, 9, 898. [Google Scholar] [CrossRef]
- Palomares-Navarro, J.J.; Bernal-Mercado, A.T.; González-Aguilar, G.A.; Ortega-Ramirez, L.A.; Martínez-Téllez, M.A.; Ayala-Zavala, J.F. Antibiofilm action of plant terpenes in Salmonella strains: Potential inhibitors of the synthesis of extracellular polymeric substances. Pathogens 2022, 12, 35. [Google Scholar] [CrossRef]
- Ju, X.; Li, J.; Zhu, M.; Lu, Z.; Lv, F.; Zhu, X.; Bie, X. Effect of the luxS gene on biofilm formation and antibiotic resistance by Salmonella serovar Dublin. Food Res. Int. 2018, 107, 385–393. [Google Scholar] [CrossRef]
- Ghosh, A.; Jayaraman, N.; Chatterji, D. Small-molecule inhibition of bacterial biofilm. ACS Omega 2020, 5, 3108–3115. [Google Scholar] [CrossRef]
- Domínguez, Á.; Muñoz, E.; López, M.C.; Cordero, M.; Martínez, J.P.; Viñas, M. Transcriptomics as a tool to discover new antibacterial targets. Biotechnol. Lett. 2017, 39, 819–828. [Google Scholar] [CrossRef]
- Dostert, M.; Trimble, M.J.; Hancock, R.E.W. Antibiofilm peptides: Overcoming biofilm-related treatment failure. RSC Adv. 2021, 11, 2718–2728. [Google Scholar] [CrossRef]
- Nazzaro, F.; Fratianni, F.; De Martino, L.; Coppola, R.; De Feo, V. Effect of essential oils on pathogenic bacteria. Pharmaceuticals 2013, 6, 1451–1474. [Google Scholar] [CrossRef]
- Guillín, Y.; Cáceres, M.; Torres, R.; Stashenko, E.; Ortiz, C. Effect of essential oils on the inhibition of biofilm and quorum sensing in Salmonella enteritidis 13076 and Salmonella typhimurium 14028. Antibiotics 2021, 10, 1191. [Google Scholar] [CrossRef] [PubMed]
- Chimnoi, N.; Reuk-Ngam, N.; Chuysinuan, P.; Khlaychan, P.; Khunnawutmanotham, N.; Chokchaichamnankit, D.; Thamniyom, W.; Klayraung, S.; Mahidol, C.; Techasakul, S. Characterization of essential oil from Ocimum gratissimum leaves: Antibacterial and mode of action against selected gastroenteritis pathogens. Microb. Pathog. 2018, 118, 290–300. [Google Scholar] [CrossRef] [PubMed]
- Seyedtaghiya, M.H.; Fasaei, B.N.; Peighambari, S.M. Antimicrobial and antibiofilm effects of Satureja hortensis essential oil against Escherichia coli and Salmonella isolated from poultry. Iran. J. Microbiol. 2021, 13, 74–80. [Google Scholar] [CrossRef] [PubMed]
- Somrani, M.; Debbabi, H.; Palop, A. Antibacterial and antibiofilm activity of essential oil of clove against Listeria monocytogenes and Salmonella enteritidis. Food Sci. Technol. Int. 2022, 28, 331–339. [Google Scholar] [CrossRef] [PubMed]
- Al-Nabulsi, A.A.; Osaili, T.M.; Olaimat, A.N.; Almasri, W.E.; Ayyash, M.; Al-Holy, M.A.; Jaradat, Z.W.; Obaid, R.S.; Holley, R.A. Inactivation of Salmonella spp. in tahini using plant essential oil extracts. Food Microbiol. 2020, 86, 103338. [Google Scholar] [CrossRef] [PubMed]
- Dutra da Silva, B.; Bernardes, P.C.; Pinheiro, P.F.; Di Giorgio Giannotti, J.; Roberto, C.D. Plectranthus amboinicus (Lour.) Spreng. essential oil as a natural alternative for the conservation of beef patties stored under refrigeration. Food Biosci. 2022, 49, 101896. [Google Scholar] [CrossRef]
- Arumugam, G.; Swamy, M.K.; Sinniah, U.R. Plectranthus amboinicus (Lour.) Spreng: Botanical, phytochemical, pharmacological and nutritional significance. Molecules 2016, 21, 369. [Google Scholar] [CrossRef]
- Leesombun, A.; Thanapakdeechaikul, K.; Suwannawiang, J.; Mukto, P.; Sungpradit, S.; Bangphoomi, N.; Changbunjong, T.; Thongjuy, O.; Weluwanarak, T.; Boonmasawai, S. Effects of Coleus amboinicus L. essential oil and ethanolic extracts on planktonic cells and biofilm formation of Microsporum canis isolated from feline dermatophytosis. Antibiotics 2022, 11, 1734. [Google Scholar] [CrossRef]
- Leesombun, A.; Sungpradit, S.; Boonmasawai, S.; Weluwanarak, T.; Klinsrithong, S.; Ruangsittichai, J.; Ampawong, S.; Masmeatathip, R.; Changbunjong, T. Insecticidal activity of Plectranthus amboinicus essential oil against the stable fly Stomoxys calcitrans (Diptera: Muscidae) and the horse fly Tabanus megalops (Diptera: Tabanidae). Insects 2022, 13, 255. [Google Scholar] [CrossRef]
- Santos, F.A.V.; Serra, C.G.; Bezerra, R.J.A.C.; Figueredo, F.G.; Edinardo; Matias, F.F.; Menezes, I.R.A.; Costa, J.G.M.; Coutinho, H.D.M. Antibacterial activity of Plectranthus amboinicus Lour (Lamiaceae) essential oil against Streptococcus mutans. Eur. J. Integr. Med. 2016, 8, 293–297. [Google Scholar] [CrossRef]
- Vasconcelos, S.E.C.B.; Melo, H.M.; Cavalcante, T.T.A.; Júnior, F.E.A.C.; de Carvalho, M.G.; Menezes, F.G.R.; de Sousa, O.V.; Costa, R.A. Plectranthus amboinicus essential oil and carvacrol bioactive against planktonic and biofilm of oxacillin- and vancomycin-resistant Staphylococcus aureus. BMC Complement. Altern. Med. 2017, 17, 462. [Google Scholar] [CrossRef] [PubMed]
- Shubha, J.R.; Bhatt, P. Plectranthus amboinicus leaves stimulate growth of probiotic L. plantarum: Evidence for ethnobotanical use in diarrhea. J. Ethnopharmacol. 2015, 166, 220–227. [Google Scholar] [CrossRef] [PubMed]
- Sarrazin, S.L.; da Silva, L.A.; de Assunção, A.P.; Oliveira, R.B.; Calao, V.Y.; da Silva, R.; Stashenko, E.E.; Maia, J.G.; Mourão, R.H. Antimicrobial and seasonal evaluation of the carvacrol-chemotype oil from Lippia origanoides kunth. Molecules 2015, 20, 1860–1871. [Google Scholar] [CrossRef] [PubMed]
- Alves Resende, J.; Soares Toneto, D.; Cruz Albuquerque, M.C.; Areas Bastos, K.; Fontes Pinheiro, P.; Drummond Costa Ignacchiti, M. Antibacterial and anti-biofilm potential of Plectranthus amboinicus (Lour.) Spreng. essential oil and carvacrol against Staphylococcus aureus and Escherichia coli. Rev. Ciências Médicas Biológicas 2022, 21, 11–17. [Google Scholar] [CrossRef]
- Monzote, L.; Scherbakov, A.M.; Scull, R.; Gutiérrez, Y.I.; Satyal, P.; Cos, P.; Shchekotikhin, A.E.; Gille, L.; Setzer, W.N. Pharmacological assessment of the carvacrol chemotype essential oil from Plectranthus amboinicus growing in Cuba. Nat. Prod. Commun. 2020, 15, 1934578X20962233. [Google Scholar]
- Trevisan, D.A.C.; Silva, A.F.; Negri, M.; Abreu Filho, B.A.; Machinski Junior, M.; Patussi, E.V.; Campanerut-Sá, P.A.Z.; Mikcha, J.M.G. Antibacterial and antibiofilm activity of carvacrol against Salmonella enterica serotype Typhimurium. Braz. J. Pharm. Sci. 2018, 54, 1–8. [Google Scholar] [CrossRef]
- Olsen, J.E.; Hoegh-Andersen, K.H.; Casadesús, J.; Rosenkranzt, J.T.; Chadfield, M.S.; Thomsen, L.E. The role of flagella and chemotaxis genes in host pathogen interaction of the host adapted Salmonella enterica serovar Dublin compared to the broad host range serovar S. Typhimurium. BMC Microbiol. 2013, 13, 67. [Google Scholar] [CrossRef]
- Soni, K.A.; Oladunjoye, A.; Nannapaneni, R.; Schilling, M.W.; Silva, J.L.; Mikel, B.; Bailey, R.H. Inhibition and inactivation of Salmonella typhimurium biofilms from polystyrene and stainless steel surfaces by essential oils and phenolic constituent carvacrol. J. Food Prot. 2013, 76, 205–212. [Google Scholar] [CrossRef]
- Burt, S.A.; Ojo-Fakunle, V.T.; Woertman, J.; Veldhuizen, E.J. The natural antimicrobial carvacrol inhibits quorum sensing in Chromobacterium violaceum and reduces bacterial biofilm formation at sub-lethal concentrations. PLoS ONE 2014, 9, e93414. [Google Scholar] [CrossRef]
- Kim, Y.K.; Roy, P.K.; Ashrafudoulla, M.; Nahar, S.; Toushik, S.H.; Hossain, M.I.; Mizan, M.F.R.; Park, S.H.; Ha, S.D. Antibiofilm effects of quercetin against Salmonella enterica biofilm formation and virulence, stress response, and quorum-sensing gene expression. Food Control. 2022, 137, 108964. [Google Scholar] [CrossRef]
- Chen, L.; Zhao, X.; Li, R.; Yang, H. Integrated metabolomics and transcriptomics reveal the adaptive responses of Salmonella enterica serovar Typhimurium to thyme and cinnamon oils. Food Res. Int. 2022, 157, 111241. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Yan, Y.; Yang, K.; Yang, X.; Dong, P.; Wu, H.; Luo, X.; Zhang, Y.; Zhu, L. Inhibitory mechanism of Salmonella Derby biofilm formation by sub-inhibitory concentrations of clove and oregano essential oil: A global transcriptomic study. Food Control. 2023, 150, 109734. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, D.F.; Zhou, X.; Xu, L.; Zhang, L.; Shi, X. Comprehensive analysis reveals two distinct evolution patterns of Salmonella flagellin gene clusters. Front. Microbiol. 2017, 8, 2604. [Google Scholar] [CrossRef]
- Wang, F.; Deng, L.; Huang, F.; Wang, Z.; Lu, Q.; Xu, C. Flagellar motility is critical for Salmonella enterica serovar Typhimurium biofilm development. Front. Microbiol. 2020, 11, 1695. [Google Scholar] [CrossRef]
- Horstmann, J.A.; Zschieschang, E.; Truschel, T.; de Diego, J.; Lunelli, M.; Rohde, M.; May, T.; Strowig, T.; Stradal, T.; Kolbe, M.; et al. Flagellin phase-dependent swimming on epithelial cell surfaces contributes to productive Salmonella gut colonisation. Cell Microbiol. 2017, 19, e12739. [Google Scholar] [CrossRef] [PubMed]
- Das, C.; Mokashi, C.; Mande, S.S.; Saini, S. Dynamics and control of flagella assembly in Salmonella typhimurium. Front. Cell Infect. Microbiol. 2018, 8, 36. [Google Scholar] [CrossRef]
- Li, B.; Yue, Y.; Yuan, Z.; Zhang, F.; Li, P.; Song, N.; Lin, W.; Liu, Y.; Yang, Y.; Li, Z.; et al. Salmonella STM1697 coordinates flagella biogenesis and virulence by restricting flagellar master protein FlhD4C2 from recruiting RNA polymerase. Nucleic Acids Res. 2017, 45, 9976–9989. [Google Scholar] [CrossRef]
- Bonifield, H.R.; Hughes, K.T. Flagellar phase variation in Salmonella enterica is mediated by a posttranscriptional control mechanism. J. Bacteriol. 2003, 185, 3567–3574. [Google Scholar] [CrossRef]
- Kolenda, R.; Ugorski, M.; Grzymajlo, K. Everything you always wanted to know about Salmonella type 1 fimbriae, but were afraid to ask. Front. Microbiol. 2019, 10, 1017. [Google Scholar] [CrossRef]
- Chen, S.; Feng, Z.; Sun, H.; Zhang, R.; Qin, T.; Peng, D. Biofilm-formation-related genes csgD and bcsA promote the vertical transmission of Salmonella enteritidis in Chicken. Front. Vet. Sci. 2020, 7, 625049. [Google Scholar] [CrossRef]
- Ogasawara, H.; Yamamoto, K.; Ishihama, A. Role of the biofilm master regulator CsgD in cross-regulation between biofilm formation and flagellar synthesis. J. Bacteriol. 2011, 193, 2587–2597. [Google Scholar] [CrossRef] [PubMed]
- Sokaribo, A.S.; Hansen, E.G.; McCarthy, M.; Desin, T.S.; Waldner, L.L.; MacKenzie, K.D.; Mutwiri, G., Jr.; Herman, N.J.; Herman, D.J.; Wang, Y.; et al. Metabolic activation of CsgD in the regulation of Salmonella biofilms. Microorganisms 2020, 8, 964. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Ma, L.; Zhao, C.; Yan, J.; Che, S.; Zhou, Z.; Wang, H.; Yang, L.; Hu, B. Transcriptome of Pectobacterium carotovorum subsp. carotovorum PccS1 infected in calla plants in vivo highlights a spatiotemporal expression pattern of genes related to virulence, adaptation, and host response. Mol. Plant Pathol. 2020, 21, 871–891. [Google Scholar] [CrossRef] [PubMed]
- Cooper, K.G.; Chong, A.; Kari, L.; Jeffrey, B.; Starr, T.; Martens, C.; McClurg, M.; Posada, V.R.; Laughlin, R.C. Whitfield-Cargile, C; et al. Regulatory protein HilD stimulates Salmonella Typhimurium invasiveness by promoting smooth swimming via the methyl-accepting chemotaxis protein McpC. Nat. Commun. 2021, 12, 348. [Google Scholar] [CrossRef]
- Ellermeier, J.R.; Slauch, J.M. Adaptation to the host environment: Regulation of the SPI1 type III secretion system in Salmonella enterica serovar Typhimurium. Curr. Opin. Microbiol. 2007, 10, 24–29. [Google Scholar] [CrossRef]
- Chaudhari, A.A.; Jasper, S.L.; Dosunmu, E.; Miller, M.E.; Arnold, R.D.; Singh, S.R.; Pillai, S. Novel pegylated silver coated carbon nanotubes kill Salmonella but they are non-toxic to eukaryotic cells. J. Nanobiotechnology 2015, 13, 23. [Google Scholar] [CrossRef]
- CLSI; Clinical and Laboratory Standards Institute. M100: Performance Standards for Antimicrobial Susceptibility Testing, 30th ed.; CLSI: Wayne, PA, USA, 2020. [Google Scholar]
- Elshikh, M.; Ahmed, S.; Funston, S.; Dunlop, P.; McGaw, M.; Marchant, R.; Banat, I.M. Resazurin-based 96-well plate microdilution method for the determination of minimum inhibitory concentration of biosurfactants. Biotechnol. Lett. 2016, 38, 1015–1019. [Google Scholar] [CrossRef]
- Mohammadi Pelarti, S.; Karimi Zarehshuran, L.; Babaeekhou, L.; Ghane, M. Antibacterial, anti-biofilm and anti-quorum sensing activities of Artemisia dracunculus essential oil (EO): A study against Salmonella enterica serovar Typhimurium and Staphylococcus aureus. Arch. Microbiol. 2021, 203, 1529–1537. [Google Scholar] [CrossRef]
- Famuyide, I.M.; Aro, A.O.; Fasina, F.O.; Eloff, J.N.; McGaw, L.J. Antibacterial and antibiofilm activity of acetone leaf extracts of nine under-investigated South African Eugenia and Syzygium (Myrtaceae) species and their selectivity indices. BMC Complement. Altern. Med. 2019, 19, 141. [Google Scholar] [CrossRef]
- Seo, M.; Oh, T.; Bae, S. Antibiofilm activity of silver nanoparticles against biofilm forming Staphylococcus pseudintermedius isolated from dogs with otitis externa. Vet. Med. Sci. 2021, 7, 1551–1557. [Google Scholar] [CrossRef]
- Audic, S.; Claverie, J.M. The significance of digital gene expression profiles. Genome. Res. 1997, 7, 986–995. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.; Islam, M.T.; Masuda, Y.; Honjoh, K.I.; Miyamoto, T. Transcriptional changes involved in inhibition of biofilm formation by ε-polylysine in Salmonella Typhimurium. Appl. Microbiol. Biotechnol. 2020, 104, 5427–5436. [Google Scholar] [CrossRef] [PubMed]










| No | Retention Time | Classes | Compounds | Formula | Chemical Structure | % of Total |
|---|---|---|---|---|---|---|
| 1 | 6.74 | Monoterpene | α-Thujene | C10H16 |  | 0.60 |
| 2 | 8.47 | Monoterpene | β-Myrcene | C10H16 |  | 1.15 |
| 3 | 8.96 | Monoterpene | α-Terpinene | C10H16 |  | 2.49 |
| 4 | 9.16 | Monoterpene | p-Cymene | C10H14 |  | 8.62 |
| 5 | 9.21 | Monoterpene | Isoterpinolene | C10H16 |  | 0.58 |
| 6 | 9.88 | Monoterpene | γ-Terpinene | C10H16 |  | 12.29 |
| 7 | 11.96 | Monoterpene | (−)-Terpinen-4-ol | C10H18O |  | 1.04 |
| 8 | 14.15 | Monoterpene | Carvacrol | C10H14O |  | 38.26 |
| 9 | 14.84 | Phenols | Eugenol | C10H12O2 |  | 0.56 |
| 10 | 15.45 | Sesquiterpene | Caryophyllene | C15H24 |  | 5.64 |
| 11 | 15.64 | Sesquiterpene | cis-α-Bisabolene | C15H24 |  | 3.71 |
| 12 | 15.88 | Sesquiterpene | Humulene | C15H24 |  | 1.54 |
| 13 | 16.72 | Fatty acids | Dodecanoic acid, methyl ester | C13H26O2 |  | 4.35 |
| 14 | 17.43 | Fatty acids | Dodecanoic acid | C12H24O2 |  | 0.73 |
| 15 | 21.04 | Fatty acids | Hexadecanoic acid, methyl ester | C17H34O2 |  | 2.27 |
| 16 | 22.62 | Fatty acids | Linoleic acid | C19H34O2 |  | 3.24 |
| 17 | 22.69 | Fatty acids | 9-Octadecenoic acid, methyl ester | C19H36O2 |  | 7.12 |
| Total | 94.17 |
| Sample Name | Total Raw Reads (Mb) | Total Clean Reads (Mb) | Total Clean Bases (Gb) | Clean Read Q20 (%) | Clean Read Q30 (%) | Clean Read Ratio (%) | GC (%) |
|---|---|---|---|---|---|---|---|
| Control | 57.94 | 55.17 | 5.52 | 98.55 | 94.94 | 95.22 | 52.17 |
| EO-CA | 45.20 | 43.16 | 4.32 | 98.65 | 95.27 | 95.49 | 52.21 |
| Gene Name | RNA-Seg Significant Fold Change (Control vs. EO-CA) | Significant Different Value | Corrected Significant Different Value | Significant (Control vs. EO-CA) |
|---|---|---|---|---|
| flhD | −1.6865 | 1.51 × 10−13 | 5.16 × 10−13 | Downregulated |
| fljB | −1.5424 | 2.01 × 10−176 | 2.37 × 10−175 | Downregulated |
| fimD | −1.9259 | 4.38 × 10−166 | 4.95 × 10−165 | Downregulated |
| adrA | −1.4871 | 2.37 × 10−155 | 2.18 × 10−114 | Downregulated |
| csgD | −0.6398 | 1.50 × 10−7 | 4.22 × 10−7 | Not significant |
| hilA | −1.5295 | 6.03 × 10−39 | 3.28 × 10−38 | Downregulated |
| Gene | Gene Function | Forward (5′−3′) | Reverse (5′−3′) |
|---|---|---|---|
| 16S rRNA | AGGCCTTCGGGTTGTAAAGT | GTTAGCCGGTGCTTCTTCTG | |
| flhD | Motility | CTCCTTGCACAGCGTTTGAT | TCTCCGCCAGTTTGACCAT |
| fljB | Motility | TGGATGTATCGGGTCTTGATG | CACCAGTAAAGCCACCAATAG |
| fimD | Motility | CGCGGCGAAAGTTATTTCAA | CCACGGACGCGGTATCC |
| adrA | Curli fimbriae | GAAGCTCGTCGCTGGAAGTC | TTCCGCTTAATTTAATGGCCG |
| csgD | Curli fimbriae | TCCTGGTCTTCAGTAGCGTAA | TATGATGGAAGCGGATAAGAA |
| hilA | Invasion | AATGGTCACAGGCTGAGGTG | ACATCGTCGCGACTTGTGAA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leesombun, A.; Sungpradit, S.; Sariya, L.; Taowan, J.; Boonmasawai, S. Transcriptional Profiling of the Effect of Coleus amboinicus L. Essential Oil against Salmonella Typhimurium Biofilm Formation. Antibiotics 2023, 12, 1598. https://doi.org/10.3390/antibiotics12111598
Leesombun A, Sungpradit S, Sariya L, Taowan J, Boonmasawai S. Transcriptional Profiling of the Effect of Coleus amboinicus L. Essential Oil against Salmonella Typhimurium Biofilm Formation. Antibiotics. 2023; 12(11):1598. https://doi.org/10.3390/antibiotics12111598
Chicago/Turabian StyleLeesombun, Arpron, Sivapong Sungpradit, Ladawan Sariya, Jarupha Taowan, and Sookruetai Boonmasawai. 2023. "Transcriptional Profiling of the Effect of Coleus amboinicus L. Essential Oil against Salmonella Typhimurium Biofilm Formation" Antibiotics 12, no. 11: 1598. https://doi.org/10.3390/antibiotics12111598
APA StyleLeesombun, A., Sungpradit, S., Sariya, L., Taowan, J., & Boonmasawai, S. (2023). Transcriptional Profiling of the Effect of Coleus amboinicus L. Essential Oil against Salmonella Typhimurium Biofilm Formation. Antibiotics, 12(11), 1598. https://doi.org/10.3390/antibiotics12111598