Subpopulations in Strains of Staphylococcus aureus Provide Antibiotic Tolerance
Abstract
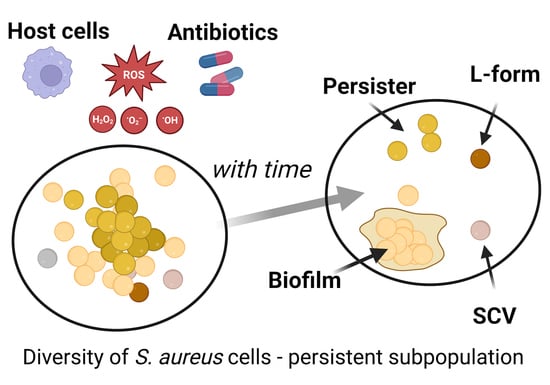
:1. Introduction
2. S. aureus and Diabetic Foot Ulcer—Foot Infection
3. Iron and S. aureus Cell Types
4. Bet-Hedging as a Model for SCV Development
5. Phenotypic Plasticity in S. aureus Persistent Populations
6. SCV and Long-Duration Antibiotic Responses
7. Genotypic and Phenotypic Changes under Antibiotic Exposure
8. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Proctor, R.A.; Kahl, B.; von Eiff, C.; Vaudaux, P.E.; Lew, D.P.; Peters, G. Staphylococcal small colony variants have novel mechanisms for antibiotic resistance. Clin. Infect. Dis. 1998, 27 (Suppl. 1), S68–S74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Proctor, R.A.; Kriegeskorte, A.; Kahl, B.C.; Becker, K.; Loffler, B.; Peters, G. Staphylococcus aureus Small Colony Variants (SCVs): A road map for the metabolic pathways involved in persistent infections. Front. Cell. Infect. Microbiol. 2014, 4, 99. [Google Scholar] [CrossRef]
- Proctor, R.A.; von Eiff, C.; Kahl, B.C.; Becker, K.; McNamara, P.; Herrmann, M.; Peters, G. Small colony variants: A pathogenic form of bacteria that facilitates persistent and recurrent infections. Nat. Rev. Microbiol. 2006, 4, 295–305. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.; Armstrong, D.G.; Lipsky, B.A. Preventing foot ulcers in patients with diabetes. JAMA 2005, 293, 217–228. [Google Scholar] [CrossRef] [PubMed]
- Messad, N.; Landraud, L.; Canivet, B.; Lina, G.; Richard, J.L.; Sotto, A.; Lavigne, J.P.; Lemichez, E. Distribution of edin in Staphylococcus aureus isolated from diabetic foot ulcers. Clin. Microbiol. Infect. 2013, 19, 875–880. [Google Scholar] [CrossRef] [Green Version]
- Sotto, A.; Lina, G.; Richard, J.-L.; Combescure, C.; Bourg, G.; Vidal, L.; Jourdan, N.; Etienne, J.; Lavigne, J.-P. Virulence Potential of Staphylococcus aureus Strains Isolated From Diabetic Foot Ulcers. Diabetes Care 2008, 31, 2318. [Google Scholar] [CrossRef] [Green Version]
- Cervantes-García, E.; García-Gonzalez, R.; Reyes-Torres, A.; Resendiz-Albor, A.A.; Salazar-Schettino, P.M. Staphylococcus aureus small colony variants in diabetic foot infections. Diabet. Foot Ankle 2015, 6, 26431. [Google Scholar] [CrossRef] [Green Version]
- Fattah, S.; Taha, A. Methicillin Resistant Small Colony Variant (Scvs) Staphylococci Isolated From Diabetic Foot Infections. Biochem. Cell. Arch. 2020, 20, 2745–2750. [Google Scholar] [CrossRef]
- Urish, K.L.; Cassat, J.E. Staphylococcus aureus Osteomyelitis: Bone, Bugs, and Surgery. Infect. Immun. 2020, 88, e00932-19. [Google Scholar] [CrossRef]
- Yang, D.; Wijenayaka, A.R.; Solomon, L.B.; Pederson, S.M.; Findlay, D.M.; Kidd, S.P.; Atkins, G.J. Novel Insights into Staphylococcus aureus Deep Bone Infections: The Involvement of Osteocytes. mBio 2018, 9, e00415-18. [Google Scholar] [CrossRef] [Green Version]
- Tuchscherr, L.; Loffler, B.; Proctor, R.A. Persistence of Staphylococcus aureus: Multiple Metabolic Pathways Impact the Expression of Virulence Factors in Small-Colony Variants (SCVs). Front. Microbiol. 2020, 11, 1028. [Google Scholar] [CrossRef]
- Painter, K.L.; Hall, A.; Ha, K.P.; Edwards, A.M. The Electron Transport Chain Sensitizes Staphylococcus aureus and Enterococcus faecalis to the Oxidative Burst. Infect. Immun. 2017, 85, e00659-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Wei, Q.; Wang, Z.; Sun, X.; He, Q.-Y. Proteomic Study of the Adaptive Mechanism of Ciprofloxacin-Resistant Staphylococcus aureus to the Host Environment. J. Proteome Res. 2022, 21, 1537–1547. [Google Scholar] [CrossRef] [PubMed]
- Friedman, D.B.; Stauff, D.L.; Pishchany, G.; Whitwell, C.W.; Torres, V.J.; Skaar, E.P. Staphylococcus aureus redirects central metabolism to increase iron availability. PLoS Pathog. 2006, 2, e87. [Google Scholar] [CrossRef] [PubMed]
- Haley, K.P.; Skaar, E.P. A battle for iron: Host sequestration and Staphylococcus aureus acquisition. Microbes Infect. 2012, 14, 217–227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hammer, N.D.; Skaar, E.P. Molecular mechanisms of Staphylococcus aureus iron acquisition. Annu. Rev. Microbiol. 2011, 65, 129–147. [Google Scholar] [CrossRef] [Green Version]
- Somerville, G.A.; Chaussee, M.S.; Morgan, C.I.; Fitzgerald, J.R.; Dorward, D.W.; Reitzer, L.J.; Musser, J.M. Staphylococcus aureus aconitase inactivation unexpectedly inhibits post-exponential-phase growth and enhances stationary-phase survival. Infect. Immun. 2002, 70, 6373–6382. [Google Scholar] [CrossRef] [Green Version]
- Somerville, G.A.; Cockayne, A.; Dürr, M.; Peschel, A.; Otto, M.; Musser, J.M. Synthesis and deformylation of Staphylococcus aureus delta-toxin are linked to tricarboxylic acid cycle activity. J. Bacteriol. 2003, 185, 6686–6694. [Google Scholar] [CrossRef] [Green Version]
- Dong, Y.; Miao, X.; Zheng, Y.-D.; Liu, J.; He, Q.-Y.; Ge, R.; Sun, X. Ciprofloxacin-Resistant Staphylococcus aureus Displays Enhanced Resistance and Virulence in Iron-Restricted Conditions. J. Proteome Res. 2021, 20, 2839–2850. [Google Scholar] [CrossRef]
- Deng, X.; Sun, F.; Ji, Q.; Liang, H.; Missiakas, D.; Lan, L.; He, C. Expression of multidrug resistance efflux pump gene norA is iron responsive in Staphylococcus aureus. J. Bacteriol. 2012, 194, 1753–1762. [Google Scholar] [CrossRef] [Green Version]
- Campion, J.J.; McNamara, P.J.; Evans, M.E. Evolution of ciprofloxacin-resistant Staphylococcus aureus in in vitro pharmacokinetic environments. Antimicrob. Agents Chemother. 2004, 48, 4733–4744. [Google Scholar] [CrossRef] [Green Version]
- Tahmasebi, H.; Dehbashi, S.; Arabestani, M.R. Antibiotic resistance alters through iron-regulating Sigma factors during the interaction of Staphylococcus aureus and Pseudomonas aeruginosa. Sci. Rep. 2021, 11, 18509. [Google Scholar] [CrossRef]
- Grimbergen, A.J.; Siebring, J.; Solopova, A.; Kuipers, O.P. Microbial bet-hedging: The power of being different. Curr. Opin. Microbiol. 2015, 25, 67–72. [Google Scholar] [CrossRef]
- Verstraete, L.; Van den Bergh, B.; Verstraeten, N.; Michiels, J. Ecology and evolution of antibiotic persistence. Trends Microbiol. 2022, 30, 466–479. [Google Scholar] [CrossRef]
- Edwards, A.M. Phenotype switching is a natural consequence of Staphylococcus aureus replication. J. Bacteriol. 2012, 194, 5404–5412. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.; Carda-Diéguez, M.; Žiemytė, M.; Vreugde, S.; Cooksley, C.; Crosby, H.A.; Horswill, A.R.; Mira, A.; Zilm, P.S.; Kidd, S.P. Functional mgrA Influences Genetic Changes within a Staphylococcus aureus Cell Population over Time. J. Bacteriol. 2022, 204, e0013822. [Google Scholar] [CrossRef]
- Von Eiff, C.; Heilmann, C.; Proctor, R.; Woltz, C.; Peters, G.; Götz, F. A Site-Directed Staphylococcus aureus hemB Mutant Is a Small Colony Variant which persists intracellularly. J. Bacteriol. 1997, 179, 4706–4712. [Google Scholar] [CrossRef] [Green Version]
- Balwit, J.M.; van Langevelde, P.; Vann, J.M.; Proctor, R. Gentamicin-resistant menadione and hemin auxotrophic Staphylococcus aureus persist within cultured endothelial cells. J. Infect. Dis. 1994, 170, 1033–1037. [Google Scholar] [CrossRef]
- Schaaff, F.; Bierbaum, G.; Baumert, N.; Bartmann, P.; Sahl, H.G. Mutations are involved in emergence of aminoglycoside-induced small colony variants of Staphylococcus aureus. Int. J. Med. Microbiol. 2003, 293, 427–435. [Google Scholar] [CrossRef]
- Lannergård, J.; Von Eiff, C.; Sander, G.; Cordes, T.; Seggewiß, J.; Peters, G.; Proctor, R.A.; Becker, K.; Hughes, D. Identification of the genetic basis for clinical menadione-auxotrophic small-colony variant isolates of Staphylococcus aureus. Antimicrob. Agents Chemother. 2008, 52, 4017–4022. [Google Scholar] [CrossRef] [Green Version]
- Pader, V.; James, E.H.; Painter, K.L.; Wigneshweraraj, S.; Edwards, A.M. The agr quorum-sensing system regulates fibronectin binding but not hemolysis in the absence of a functional electron transport chain. Infect. Immun. 2014, 82, 4337–4347. [Google Scholar] [CrossRef] [Green Version]
- Dean, M.A.; Olsen, R.J.; Wesley Long, S.; Rosato, A.E.; Musser, J.M. Identification of point mutations in clinical Staphylococcus aureus strains that produce small-colony variants auxotrophic for menadione. Infect. Immun. 2014, 82, 1600–1605. [Google Scholar] [CrossRef] [Green Version]
- Kittinger, C.; Toplitsch, D.; Folli, B.; Masoud Landgraf, L.; Zarfel, G. Phenotypic Stability of Staphylococcus aureus Small Colony Variants (SCV) Isolates from Cystic Fibrosis (CF) Patients. Int. J. Environ. Res. Public Health 2019, 16, 1940. [Google Scholar] [CrossRef] [Green Version]
- Chatterjee, I.; Kriegeskorte, A.; Fischer, A.; Deiwick, S.; Theimann, N.; Proctor, R.A.; Peters, G.; Herrmann, M.; Kahl, B.C. In vivo mutations of thymidylate synthase (Encoded by thyA) are responsible for thymidine dependency in clinical small-colony variants of Staphylococcus aureus. J. Bacteriol. 2008, 190, 834–842. [Google Scholar] [CrossRef] [Green Version]
- Besier, S.; Ludwig, A.; Ohlsen, K.; Brade, V.; Wichelhaus, T.A. Molecular analysis of the thymidine-auxotrophic small colony variant phenotype of Staphylococcus aureus. Int. J. Med. Microbiol. 2007, 297, 217–225. [Google Scholar] [CrossRef]
- Gómez-González, C.; Acosta, J.; Villa, J.; Barrado, L.; Sanz, F.; Orellana, M.Á.; Otero, J.R.; Chaves, F. Clinical and molecular characteristics of infections with CO2-dependent small-colony variants of Staphylococcus aureus. J. Clin. Microbiol. 2010, 48, 2878–2884. [Google Scholar] [CrossRef] [Green Version]
- Thomas, M.E. Studies on a CO2-dependent staphylococcus. J. Clin. Pathol. 1955, 8, 288–291. [Google Scholar] [CrossRef] [Green Version]
- Bazaid, A.S.; Forbes, S.; Humphreys, G.J.; Ledder, R.G.; O’Cualain, R.; McBain, A.J. Fatty Acid Supplementation Reverses the Small Colony Variant Phenotype in Triclosan-Adapted Staphylococcus aureus: Genetic, Proteomic and Phenotypic Analyses. Sci. Rep. 2018, 8, 3876. [Google Scholar] [CrossRef] [Green Version]
- Schleimer, N.; Kaspar, U.; Drescher, M.; Seggewiß, J.; von Eiff, C.; Proctor, R.A.; Peters, G.; Kriegeskorte, A.; Becker, K. The energy-coupling factor transporter module EcfAA’T, a novel candidate for the genetic basis of fatty acid-auxotrophic small-colony variants of Staphylococcus aureus. Front. Microbiol. 2018, 9, 1863. [Google Scholar] [CrossRef] [Green Version]
- Zhang, P.; Wright, J.A.; Osman, A.A.; Nair, S.P. An aroD Ochre mutation results in a Staphylococcus aureus small colony variant that can undergo phenotypic switching via two alternative mechanisms. Front. Microbiol. 2017, 8, 1001. [Google Scholar] [CrossRef] [Green Version]
- Mitchell, G.; Fugere, A.; Pepin Gaudreau, K.; Brouillette, E.; Frost, E.H.; Cantin, A.M.; Malouin, F. SigB is a dominant regulator of virulence in Staphylococcus aureus small-colony variants. PLoS ONE 2013, 8, e65018. [Google Scholar] [CrossRef] [Green Version]
- Bui, L.M.G.; Kidd, S.P. A full genomic characterization of the development of a stable Small Colony Variant cell-type by a clinical Staphylococcus aureus strain. Infect. Genet. Evol. 2015, 36, 345–355. [Google Scholar] [CrossRef]
- Favate, J.S.; Liang, S.; Cope, A.L.; Yadavalli, S.S.; Shah, P. The landscape of transcriptional and 1translational changes over 22 years of bacterial adaptation. Elife 2022, 11, e81979. [Google Scholar] [CrossRef]
- Girgis, H.S.; Harris, K.; Tavazoie, S. Large mutational target size for rapid emergence of bacterial persistence. Proc. Natl. Acad. Sci. USA 2012, 109, 12740–12745. [Google Scholar] [CrossRef] [Green Version]
- Guerillot, R.; Kostoulias, X.; Donovan, L.; Li, L.; Carter, G.P.; Hachani, A.; Vandelannoote, K.; Giulieri, S.; Monk, I.R.; Kunimoto, M.; et al. Unstable chromosome rearrangements in Staphylococcus aureus cause phenotype switching associated with persistent infections. Proc. Natl. Acad. Sci. USA 2019, 116, 20135–20140. [Google Scholar] [CrossRef] [Green Version]
- Bui, L.M.G.; Hoffmann, P.; Turnidge, J.D.; Zilm, P.S.; Kidd, S.P. Prolonged growth of a clinical Staphylococcus aureus strain selects for a stable small-colony-variant cell type. Infect. Immun. 2015, 83, 470–481. [Google Scholar] [CrossRef] [Green Version]
- Tuchscherr, L.; Buzzola, F.R.; Alvarez, L.P.; Lee, J.C.; Sordelli, D.O. Antibodies to capsular polysaccharide and clumping factor a prevent mastitis and the emergence of unencapsulated and small-colony variants of Staphylococcus aureus in mice. Infect. Immun. 2008, 76, 5738–5744. [Google Scholar] [CrossRef] [Green Version]
- Kahl, B.C.; Duebbers, A.; Lubritz, G.; Haeberle, J.; Koch, H.G.; Ritzerfeld, B.; Reilly, M.; Harms, E.; Proctor, R.A.; Herrmann, M.; et al. Population dynamics of persistent Staphylococcus aureus isolated from the airways of cystic fibrosis patients during a 6-year prospective study. J. Clin. Microbiol. 2003, 41, 4424–4427. [Google Scholar] [CrossRef] [Green Version]
- Cao, S.; Huseby, D.L.; Brandis, G.; Hughes, D. Alternative Evolutionary Pathways for Drug-Resistant Small Colony Variant Mutants in Staphylococcus aureus. mBio 2017, 8, e00358-17. [Google Scholar] [CrossRef] [Green Version]
- Kinkel, T.L.; Roux, C.M.; Dunman, P.M.; Fang, F.C. The Staphylococcus aureus SrrAB two-component system promotes resistance to nitrosative stress and hypoxia. mBio 2013, 4, e00696-13. [Google Scholar] [CrossRef] [Green Version]
- Banville, R.R. Factors affecting growth of Staphylococcus aureus L forms on semidefined medium. J. Bacteriol. 1964, 87, 1192–1197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Falcon, M.A.; Mansito, T.B.; Carnicero, A.; Gutierrez-Navarro, A.M. L-form-like colonies of Staphylococcus aureus induced by an extracellular lytic enzyme from Pseudomonas aeruginosa. J. Clin. Microbiol. 1989, 27, 1650–1654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Owens, W.E.; Nickerson, S.C. Morphologic study of Staphylococcus aureus L-form, reverting, and intermediate colonies in situ. J. Clin. Microbiol. 1989, 27, 1382–1386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quie, P.G. Microcolonies (G-variants) of Staphylococcus aureus. Yale J. Biol. Med. 1969, 41, 394–403. [Google Scholar]
- Yabu, K. Conversion of Staphylococcus aureus cells to stable L-forms in a liquid growth medium. Microbiol. Immunol. 1985, 29, 157–161. [Google Scholar] [CrossRef]
- Xu, Y.; Zhang, B.; Wang, L.; Jing, T.; Chen, J.; Xu, X.; Zhang, W.; Zhang, Y.; Han, J. Unusual features and molecular pathways of Staphylococcus aureus L-form bacteria. Microb. Pathog. 2020, 140, 103970. [Google Scholar] [CrossRef]
- Kawai, Y.; Mercier, R.; Mickiewicz, K.; Serafini, A.; Sório de Carvalho, L.P.; Errington, J. Crucial role for central carbon metabolism in the bacterial L-form switch and killing by β-lactam antibiotics. Nat. Microbiol. 2019, 4, 1716–1726. [Google Scholar] [CrossRef]
- Peyrusson, F.; Nguyen, T.K.; Najdovski, T.; Van Bambeke, F. Host Cell Oxidative Stress Induces Dormant Staphylococcus aureus Persisters. Microbiol. Spectr. 2022, 10, e0231321. [Google Scholar] [CrossRef]
- Otto, M. Staphylococcal Biofilms. Microbiol. Spectr. 2018, 6, 4. [Google Scholar] [CrossRef]
- Schilcher, K.; Horswill, A.R. Staphylococcal Biofilm Development: Structure, Regulation, and Treatment Strategies. Microbiol. Mol. Biol. Rev. 2020, 84, e00026-19. [Google Scholar] [CrossRef]
- Bui, L.M.; Turnidge, J.D.; Kidd, S.P. The induction of Staphylococcus aureus biofilm formation or Small Colony Variants is strain specific response to host-generated chemical stresses. Microbes Infect. 2015, 17, 77–82. [Google Scholar] [CrossRef] [PubMed]
- Gresham, D.; Hong, J. The functional basis of adaptive evolution in chemostats. FEMS Microbiol. Rev. 2014, 39, 2–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fait, A.; Seif, Y.; Mikkelsen, K.; Poudel, S.; Wells, J.M.; Palsson, B.O.; Ingmer, H. Adaptive laboratory evolution and independent component analysis disentangle complex vancomycin adaptation trajectories. Proc. Natl. Acad. Sci. USA 2022, 119, e2118262119. [Google Scholar] [CrossRef] [PubMed]
- Fridman, O.; Goldberg, A.; Ronin, I.; Shoresh, N.; Balaban, N.Q. Optimization of lag time underlies antibiotic tolerance in evolved bacterial populations. Nature 2014, 513, 418–421. [Google Scholar] [CrossRef]
- Kotrba, P.; Inui, M.; Yukawa, H. Bacterial phosphotransferase system (PTS) in carbohydrate uptake and control of carbon metabolism. J. Biosci. Bioeng. 2001, 92, 502–517. [Google Scholar] [CrossRef]
- Torres, V.J.; Attia, A.S.; Mason, W.J.; Hood, M.I.; Corbin, B.D.; Beasley, F.C.; Anderson, K.L.; Stauff, D.L.; McDonald, W.H.; Zimmerman, L.J.; et al. Staphylococcus aureus fur regulates the expression of virulence factors that contribute to the pathogenesis of pneumonia. Infect. Immun. 2010, 78, 1618–1628. [Google Scholar] [CrossRef] [Green Version]
- Méhi, O.; Bogos, B.; Csörgő, B.; Pál, F.; Nyerges, A.; Papp, B.; Pál, C. Perturbation of iron homeostasis promotes the evolution of antibiotic resistance. Mol. Biol. Evol. 2014, 31, 2793–2804. [Google Scholar] [CrossRef] [Green Version]
- Oliver, A.; Levin, B.R.; Juan, C.; Baquero, F.; Blázquez, J. Hypermutation and the Preexistence of Antibiotic-Resistant Pseudomonas aeruginosa Mutants: Implications for Susceptibility Testing and Treatment of Chronic Infections. Antimicrob. Agents Chemother. 2004, 48, 4226–4233. [Google Scholar] [CrossRef] [Green Version]
- Vitko, N.P.; Grosser, M.R.; Khatri, D.; Lance, T.R.; Richardson, A.R. Expanded glucose import capability affords Staphylococcus aureus optimized glycolytic flux during infection. mBio 2016, 7, e00296-16. [Google Scholar] [CrossRef] [Green Version]
- Hussain, M.; Hastings, J.G.M.; White, P.J. A chemically defined medium for slime production by coagulase-negative staphylococci. J. Med. Microbiol. 1991, 34, 143–147. [Google Scholar] [CrossRef] [Green Version]
- Jahn, L.J.; Munck, C.; Ellabaan, M.M.H.; Sommer, M.O.A. Adaptive Laboratory Evolution of Antibiotic Resistance Using Different Selection Regimes Lead to Similar Phenotypes and Genotypes. Front. Microbiol. 2017, 8, 816. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Cell Type | Phenotypic Changes | Genotypic Changes | Metabolic Changes |
|---|---|---|---|
| SCV | Small colony size (<10th normal size) Altered or no pigment Stable SCV Reverting SCV | SNP 2 (various genes) Genomic duplications Genomic inversions Gene deletions | Slow growth Low intracellular ATP (defective electron transport chain) Increased intracellular ROS 1 |
| L-forms | Reduced or lack of peptidoglycan Slow replication Stable and reverting forms. | To be determined (for S. aureus) | Increased respiratory chain Increased intracellular ROS Downregulation of virulence factors Increased glycolysis |
| Persisters | Antibiotic tolerance | Transcriptional changes SNPs (various genes) | Low intracellular ATP Increased intracellular ROS Damaged TCA |
| Biofilm cells | Surface structures/proteins Cell–cell interactions Extracellular polymeric substance (EPS) | Complex arrays of transcriptional changes (mainly) Quorum Sensing | The population of cells in a biofilm include persister cells |
| UA-DI-55 | |
|---|---|
| Colony type | Large |
| Non-pigmented | |
| Stable | |
| Genotype | MRSA |
| Patient background: | |
| Age | 80 yo |
| Sex | Female |
| Condition | Diabetes (10 y) |
| Infection | Osteomyelitis |
| Source | Intracellular bone |
| Strain | Ciprofloxacin | Penicillin | Gentamycin | |||
|---|---|---|---|---|---|---|
| MIC 1 | MBC | MIC | MBC | MIC | MBC | |
| UA-DI-55 | 250 | 750 | 250 | 750 | 62 | 750 |
| UA-DI-55-d7 | >1000 | >1000 | >1000 | >1000 | >1000 | >1000 |
| UA-DI-55-d20 | >1000 | >1000 | >1000 | >1000 | >1000 | >1000 |
| Gene | Nucleotide Change | Amino Acid Change | Product |
|---|---|---|---|
| fur | 20 (G-C) | Arg7Pro | Ferric uptake repressor |
| hypothetical | 475 (A-G) | Lys154Glu | - |
| ptsH | 236_238 (dupACG) 1 | Asp79dup | Phospho-carrier protein |
| hypothetical | 548 (A-T) | Glu183Val | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mashayamombe, M.; Carda-Diéguez, M.; Mira, A.; Fitridge, R.; Zilm, P.S.; Kidd, S.P. Subpopulations in Strains of Staphylococcus aureus Provide Antibiotic Tolerance. Antibiotics 2023, 12, 406. https://doi.org/10.3390/antibiotics12020406
Mashayamombe M, Carda-Diéguez M, Mira A, Fitridge R, Zilm PS, Kidd SP. Subpopulations in Strains of Staphylococcus aureus Provide Antibiotic Tolerance. Antibiotics. 2023; 12(2):406. https://doi.org/10.3390/antibiotics12020406
Chicago/Turabian StyleMashayamombe, Matipaishe, Miguel Carda-Diéguez, Alex Mira, Robert Fitridge, Peter S. Zilm, and Stephen P. Kidd. 2023. "Subpopulations in Strains of Staphylococcus aureus Provide Antibiotic Tolerance" Antibiotics 12, no. 2: 406. https://doi.org/10.3390/antibiotics12020406
APA StyleMashayamombe, M., Carda-Diéguez, M., Mira, A., Fitridge, R., Zilm, P. S., & Kidd, S. P. (2023). Subpopulations in Strains of Staphylococcus aureus Provide Antibiotic Tolerance. Antibiotics, 12(2), 406. https://doi.org/10.3390/antibiotics12020406