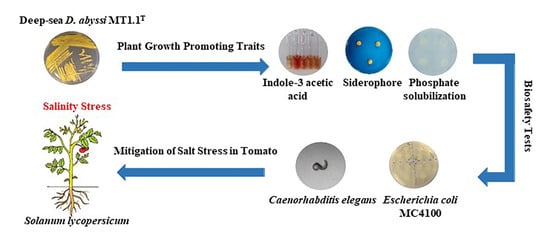
Plant Beneficial Deep-Sea Actinobacterium, Dermacoccus abyssi MT1.1T Promote Growth of Tomato (Solanum lycopersicum) under Salinity Stress
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
2.2. Assessment of Plant Growth Promoting Traits of Actinobacteria
2.3. Genomic Analysis for Plant Growth-Promoting Properties
2.4. Promotion of Tomato Growth by D. abyssi MT1.1T under Salt Stress Condition
2.4.1. Plant Growth Condition and Bacterial Inoculation
2.4.2. Root Colonization by D. abyssi MT1.1T
2.5. Biosafety Test for Actinobacteria
2.5.1. Pathogenicity Bioassay Based on Caenorhabditis elegans
2.5.2. Escherichia coli MC4100 Sensitivity
2.6. Statistical Analysis
3. Results
3.1. Assessment of Plant Growth Promoting Traits of Actinobacteria
3.2. Genomic Analysis for Plant Growth-Promoting Properties and Stress Response
3.3. Promotion of Tomato Growth by D. abyssi MT1.1T under Salt Stress Condition
Tomato Root Colonization by D. abyssi MT1.1T
3.4. Biosafety Test for Actinobacteria
3.4.1. Pathogenicity Bioassay Based on Caenorhabditis elegans
3.4.2. Escherichia coli MC4100 Sensitivity
4. Discussion
4.1. Assessment of Plant Growth Promoting Abilities of Actinobacteria
4.2. Genomic Analysis for Plant Growth-Promoting Properties
4.3. Promotion of Tomato Growth by D. abyssi MT1.1T under Salt Stress Condition
4.4. Biosafety Test for Actinobacteria
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Shrivastava, P.; Kumar, R. Soil salinity : A serious environmental issue and plant growth promoting bacteria as one of the tools for its alleviation. Saudi J. Biol. Sci. 2015, 22, 123–131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Butcher, K.; Wick, A.F.; Desutter, T.; Chatterjee, A.; Harmon, J. Soil salinity: A threat to global food security. Agron. J. 2016, 108, 2189–2200. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Nahar, K.; Alam, M.M.; Bhowmik, P.C.; Hossain, M.A.; Rahman, M.M.; Prasad, M.N.V.; Ozturk, M.; Fujita, M. Potential use of halophytes to remediate saline soils. Biomed Res. Int. 2014, 2014, 589341. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Vinocur, B.; Altman, A. Plant responses to drought, salinity and extreme temperatures: Towards genetic engineering for stress tolerance. Planta 2003, 218, 1–14. [Google Scholar] [CrossRef]
- Parihar, P.; Singh, S.; Singh, R.; Singh, V.; Prasad, S. Effect of salinity stress on plants and its tolerance strategies: A review. Environ. Sci. Pollut. Res. 2015, 22, 4056–4075. [Google Scholar] [CrossRef]
- Javed, Q.; Azeem, A.; Jabran, K.; Kumar, A. Impacts of salt stress on the physiology of plants and opportunity to rewater the stressed plants with diluted water: A review. Appl. Ecol. Environ. Res. 2019, 17, 12583–12604. [Google Scholar] [CrossRef]
- Reza Yousefi, A.; Rashidi, S.; Moradi, P.; Mastinu, A. Germination and Seedling Growth Responses of Zygophyllum fabago, Salsola kali L. and Atriplex canescens to PEG-Induced Drought Stress. Environments 2020, 7, 107. [Google Scholar] [CrossRef]
- Acosta-Motos, J.; Ortuño, M.; Bernal-Vicente, A.; Diaz-Vivancos, P.; Sanchez-Blanco, M.; Hernandez, J. Plant responses to salt stress: Adaptive mechanisms. Agronomy 2017, 7, 18. [Google Scholar] [CrossRef] [Green Version]
- Hanin, M.; Ebel, C.; Ngom, M.; Laplaze, L.; Masmoudi, K. New insights on plant salt tolerance mechanisms and their potential use for breeding. Front. Plant Sci. 2016, 7, 1787. [Google Scholar] [CrossRef] [Green Version]
- Munns, R.; Tester, M. Mechanisms of Salinity Tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [Green Version]
- Kamran, M.; Parveen, A.; Ahmar, S.; Hussain, S.; Chattha, M.S.; Saleem, M.H.; Adil, M.; Heidari, P.; Chen, J. An overview of hazardous impacts of soil salinity in crops, tolerance mechanisms, and amelioration through selenium supplementation. Int. J. Mol. Sci. 2020, 21, 148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Numan, M.; Bashir, S.; Khan, Y.; Mumtaz, R.; Shinwari, Z.K.; Khan, A.L.; Khan, A.; AL-Harrasi, A. Plant growth promoting bacteria as an alternative strategy for salt tolerance in plants: A review. Microbiol. Res. 2018, 209, 21–32. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Jha, A.B.; Dubey, R.S.; Pessarakli, M. Reactive Oxygen Species, Oxidative Damage, and Antioxidative Defense Mechanism in Plants under Stressful Conditions. J. Bot. 2012, 2012, 217037. [Google Scholar] [CrossRef] [Green Version]
- Palaniyandi, S.A.; Damodharan, K.; Yang, S.H.; Suh, J.W. Streptomyces sp. strain PGPA39 alleviates salt stress and promotes growth of “Micro Tom” tomato plants. J. Appl. Microbiol. 2014, 117, 766–773. [Google Scholar] [CrossRef] [PubMed]
- Gong, Y.; Chen, L.J.; Pan, S.Y.; Li, X.W.; Xu, M.J.; Zhang, C.M.; Xing, K.; Qin, S. Antifungal potential evaluation and alleviation of salt stress in tomato seedlings by a halotolerant plant growth-promoting actinomycete Streptomyces sp. KLBMP5084. Rhizosphere 2020, 16, 100262. [Google Scholar] [CrossRef]
- Xiong, Y.W.; Gong, Y.; Li, X.W.; Chen, P.; Ju, X.Y.; Zhang, C.M.; Yuan, B.; Lv, Z.P.; Xing, K.; Qin, S. Enhancement of growth and salt tolerance of tomato seedlings by a natural halotolerant actinobacterium Glutamicibacter halophytocola KLBMP 5180 isolated from a coastal halophyte. Plant Soil 2019, 445, 307–322. [Google Scholar] [CrossRef]
- Rangseekaew, P.; Barros-Rodríguez, A.; Pathom-Aree, W.; Manzanera, M. Deep-sea actinobacteria mitigate salinity stress in tomato seedlings and their biosafety testing. Plants 2021, 10, 1687. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.A.; Ullah, I.; Waqas, M.; Hamayun, M.; Khan, A.L.; Asaf, S.; Kang, S.; Kim, K.; Jan, R.; Lee, I. Halo-tolerant rhizospheric Arthrobacter woluwensis AK1 mitigates salt stress and induces physio-hormonal changes and expression of GmST1 and GmLAX3 in soybean. Symbiosys 2019, 77, 9–21. [Google Scholar] [CrossRef]
- Khan, M.A.; Sahile, A.A.; Jan, R.; Asaf, S.; Hamayun, M.; Imran, M.; Adhikari, A.; Kang, S.; Kim, K.; Lee, I. Halotolerant bacteria mitigate the effects of salinity stress on soybean growth by regulating secondary metabolites and molecular responses. BMC Plant Biol. 2021, 21, 176. [Google Scholar] [CrossRef]
- Sadeghi, A.; Karimi, E.; Dahaji, P.A.; Javid, M.G.; Dalvand, Y.; Askari, H. Plant growth promoting activity of an auxin and siderophore producing isolate of Streptomyces under saline soil conditions. World J. Microbiol. Biotechnol. 2012, 28, 1503–1509. [Google Scholar] [CrossRef]
- Djebaili, R.; Pellegrini, M.; Rossi, M.; Forni, C.; Smati, M.; Del Gallo, M.; Kitouni, M. Characterization of plant growth-promoting traits and inoculation effects on triticum durum of actinomycetes isolates under salt stress conditions. Soil Syst. 2021, 5, 26. [Google Scholar] [CrossRef]
- Saidi, S.; Cherif, H.; Ali, S.; Bouket, C.; Silini, A.; Eshelli, M.; Luptakova, L. Improvement of Medicago sativa Crops Productivity by the Co-inoculation of Sinorhizobium meliloti–Actinobacteria Under Salt Stress. Curr. Microbiol. 2021, 78, 1344–1357. [Google Scholar] [CrossRef]
- Mathew, B.T.; Torky, Y.; Amin, A.; Mourad, A.H.I.; Ayyash, M.M.; El-Keblawy, A.; Hilal-Alnaqbi, A.; AbuQamar, S.F.; El-Tarabily, K.A. Halotolerant Marine Rhizosphere-Competent Actinobacteria Promote Salicornia bigelovii Growth and Seed Production Using Seawater Irrigation. Front. Microbiol. 2020, 11, 552. [Google Scholar] [CrossRef] [PubMed]
- Suksaard, P.; Pathom-aree, W.; Duangmal, K. Diversity and plant growth promoting activities of actinomycetes from mangroves. Chiang Mai J. Sci. 2017, 44, 1210–1223. [Google Scholar]
- Sathya, A.; Vijayabharathi, R.; Gopalakrishnan, S. Plant growth-promoting actinobacteria: A new strategy for enhancing sustainable production and protection of grain legumes. 3 Biotech 2017, 7, 102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Glick, B.R. Plant growth-promoting bacteria: Mechanisms and applications. Scientifica 2012, 2012, 963401. [Google Scholar] [CrossRef] [Green Version]
- Sagar, A.; Rai, S.; Ilyas, N.; Sayyed, R.Z.; Al-Turki, A.I.; El Enshasy, H.A.; Simarmata, T. Halotolerant Rhizobacteria for Salinity-Stress Mitigation: Diversity, Mechanisms and Molecular Approaches. Sustainability 2022, 14, 490. [Google Scholar] [CrossRef]
- Vílchez, J.I.; Niehaus, K.; Dowling, D.N.; González-López, J.; Manzanera, M. Protection of Pepper Plants from Drought by Microbacterium sp. 3J1 by Modulation of the Plant’s Glutamine and α-ketoglutarate Content: A Comparative Metabolomics Approach. Front. Microbiol. 2018, 9, 284. [Google Scholar] [CrossRef]
- Sharma, A.; Shahzad, B.; Kumar, V.; Kohli, S.K.; Sidhu, G.P.S.; Bali, A.S.; Handa, N.; Kapoor, D.; Bhardwaj, R.; Zheng, B. Phytohormones regulate accumulation of osmolytes under abiotic stress. Biomolecules 2019, 9, 285. [Google Scholar] [CrossRef] [Green Version]
- Sofo, A.; Scopa, A.; Nuzzaci, M.; Vitti, A. Ascorbate peroxidase and catalase activities and their genetic regulation in plants subjected to drought and salinity stresses. Int. J. Mol. Sci. 2015, 16, 13561–13578. [Google Scholar] [CrossRef] [Green Version]
- Kumar, A.; Singh, S.; Gaurav, A.K.; Srivastava, S. Plant Growth-Promoting Bacteria : Biological Tools for the Mitigation of Salinity Stress in Plants. Front. Microbiol. 2020, 11, 1216. [Google Scholar] [CrossRef] [PubMed]
- Orozco-mosqueda, C.; Glick, B.R.; Santoyo, G. ACC deaminase in plant growth-promoting bacteria (PGPB): An e ffi cient mechanism to counter salt stress in crops. Microbiol. Res. 2020, 235, 126439. [Google Scholar] [CrossRef]
- Barros-Rodríguez, A.; Rangseekaew, P.; Lasudee, K.; Pathom-aree, W.; Manzanera, M. Regulatory risks associated with bacteria as biostimulants and biofertilizers in the frame of the European Regulation (EU) 2019/1009. Sci. Total Environ. 2020, 740, 140239. [Google Scholar] [CrossRef] [PubMed]
- Pathom-Aree, W.; Nogi, Y.; Sutcliffe, I.C.; Ward, A.C.; Horikoshi, K.; Bull, A.T.; Goodfellow, M. Dermacoccus abyssi sp. nov., a piezotolerant actinomycete isolated from the Mariana Trench. Int. J. Syst. Evol. Microbiol. 2006, 56, 1233–1237. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Mageed, W.M.; Juhasz, B.; Lehri, B.; Alqahtani, A.S.; Nouioui, I.; Pech-Puch, D.; Tabudravu, J.N.; Goodfellow, M.; Rodríguez, J.; Jaspars, M.; et al. Whole genome sequence of dermacoccus abyssi MT1.1 isolated from the challenger deep of the mariana trench reveals phenazine biosynthesis locus and environmental adaptation factors. Mar. Drugs 2020, 18, 131. [Google Scholar] [CrossRef] [Green Version]
- Pathom-aree, W.; Nogi, Y.; Ward, A.C.; Horikoshi, K.; Bull, A.T.; Goodfellow, M. Dermacoccus barathri sp. nov. and Dermacoccus profundi sp. nov., novel actinomycetes isolated from deep-sea mud of the Mariana Trench. Int. J. Syst. Evol. Microbiol. 2006, 56, 2303–2307. [Google Scholar] [CrossRef]
- Lasudee, K.; Tokuyama, S.; Lumyong, S.; Pathom-Aree, W. Actinobacteria Associated with arbuscular mycorrhizal funneliformis mosseae spores, taxonomic characterization and their beneficial traits to plants: Evidence obtained from mung bean (Vigna radiata) and Thai Jasmine Rice (Oryza sativa). Front. Microbiol. 2018, 9, 1247. [Google Scholar] [CrossRef] [PubMed]
- Schwyn, B.; Neilands, J.B. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 1987, 160, 47–56. [Google Scholar] [CrossRef]
- Djebaili, R.; Pellegrini, M.; Smati, M.; Del Gallo, M.; Kitouni, M. Actinomycete strains isolated from saline soils: Plant-growth-promoting traits and inoculation effects on solanum lycopersicum. Sustainability 2020, 12, 4617. [Google Scholar] [CrossRef]
- Overbeek, R.; Olson, R.; Pusch, G.D.; Olsen, G.J.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Parrello, B.; Shukla, M.; et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 2014, 42, 206–214. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Kloosterman, A.M.; Charlop-Powers, Z.; van Wezel, G.P.; Medema, M.H.; Weber, T. AntiSMASH 6.0: Improving cluster detection and comparison capabilities. Nucleic Acids Res. 2021, 49, W29–W35. [Google Scholar] [CrossRef] [PubMed]
- Skinnider, M.A.; Merwin, N.J.; Johnston, C.W.; Magarvey, N.A. PRISM 3: Expanded prediction of natural product chemical structures from microbial genomes. Nucleic Acids Res. 2017, 45, W49–W54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cosentino, S.; Voldby Larsen, M.; Møller Aarestrup, F.; Lund, O. PathogenFinder - Distinguishing friend from foe using bacterial whole genome sequence data. PLoS ONE 2013, 8, e77302. [Google Scholar] [CrossRef]
- Narváez-Reinaldo, J.J.; Barba, I.; González-López, J.; Tunnacliffe, A.; Manzanera, M. Rapid Method for Isolation of Desiccation-Tolerant Strains and Xeroprotectants. Appl. Environ. Microbiol. 2010, 76, 5254–5262. [Google Scholar] [CrossRef] [Green Version]
- Vílchez, J.I.; García-Fontana, C.; Román-Naranjo, D.; González-López, J.; Manzanera, M. Plant drought tolerance enhancement by trehalose production of desiccation-tolerant microorganisms. Front. Microbiol. 2016, 7, 1577. [Google Scholar] [CrossRef] [Green Version]
- Oukarroum, A.; El Madidi, S.; Schansker, G.; Strasser, R.J. Probing the responses of barley cultivars (Hordeum vulgare L.) by chlorophyll a fluorescence OLKJIP under drought stress and re-watering. Environ. Exp. Bot. 2007, 60, 438–446. [Google Scholar] [CrossRef]
- Ashraf, M.; Shahzad, S.M.; Akhtar, N.; Imtiaz, M.; Ali, A. Salinization/sodification of soil and physiological dynamics of sunflower irrigated with saline–sodic water amending by potassium and farm yard manure. J. Water Reuse Desalin. 2017, 7, 476–487. [Google Scholar] [CrossRef]
- Arnon, D.I. Copper enzymes in isolated chloroplasts. polyphenoloxidase in Beta vulgaris. Plant Physiol. 1949, 24, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Bates, L.S.; Waldren, R.P.; Teare, I.D. Rapid determination of free proline for water-stress studies. Plant Soil 1973, 39, 205–207. [Google Scholar] [CrossRef]
- DuBois, M.; Gilles, K.A.; Hamilton, J.K.; Rebers, P.A.; Smith, F. Colorimetric method for determination of sugars and related substances. Anal. Chem. 1956, 28, 350–356. [Google Scholar] [CrossRef]
- Velikova, V.; Yordanov, I.; Edreva, A. Oxidative stress and some antioxidant systems in acid rain-treated bean plants. Plant Sci. 2000, 151, 59–66. [Google Scholar] [CrossRef]
- Botta, A.L.; Santacecilia, A.; Ercole, C.; Cacchio, P.; Del Gallo, M. In vitro and in vivo inoculation of four endophytic bacteria on Lycopersicon esculentum. N. Biotechnol. 2013, 30, 666–674. [Google Scholar] [CrossRef] [PubMed]
- Vílchez, J.I.; Navas, A.; González-lópez, J.; Arcos, S.C.; Gutierrez, F.J. Biosafety test for plant growth-promoting bacteria: Proposed Environmental and Human Safety Index (EHSI) protocol. Front. Microbiol. 2016, 6, 1514. [Google Scholar] [CrossRef] [PubMed]
- Vílchez, S.; Tunnacliffe, A.; Manzanera, M. Tolerance of plastic-encapsulated Pseudomonas putida KT2440 to chemical stress. Extremophiles 2008, 12, 297–299. [Google Scholar] [CrossRef] [PubMed]
- García-Fontana, C.; Vílchez, J.I.; González-Requena, M.; González-López, J.; Krell, T.; Matilla, M.A.; Manzanera, M. The involvement of McpB chemoreceptor from Pseudomonas aeruginosa PAO1 in virulence. Sci. Rep. 2019, 9, 13166. [Google Scholar] [CrossRef]
- Hoelzle, K.; Peter, S.; Sidler, M.; Kramer, M.M.; Wittenbrink, M.M.; Felder, K.M.; Hoelzle, L.E. Inorganic pyrophosphatase in uncultivable hemotrophic mycoplasmas: Identification and properties of the enzyme from Mycoplasma suis. BMC Microbiol. 2010, 10, 194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhatti, A.A.; Haq, S.; Bhat, R.A. Actinomycetes benefaction role in soil and plant health. Microb. Pathog. 2017, 111, 458–467. [Google Scholar] [CrossRef] [PubMed]
- Tsavkelova, E.A.; Klimova, S.Y.; Cherdyntseva, T.A.; Netrusov, A.I. Microbial producers of plant growth stimulators and their practical use: A review. Appl. Biochem. Microbiol. 2006, 42, 117–126. [Google Scholar] [CrossRef]
- Wang, W.; Qiu, Z.; Tan, H.; Cao, L. Siderophore production by actinobacteria. BioMetals 2014, 27, 623–631. [Google Scholar] [CrossRef]
- Alori, E.T.; Glick, B.R.; Babalola, O.O. Microbial phosphorus solubilization and its potential for use in sustainable agriculture. Front. Microbiol. 2017, 8, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Zhu, F.; Qu, L.; Hong, X.; Sun, X. Isolation and characterization of a phosphate- solubilizing halophilic bacterium Kushneria sp. YCWA18 from Daqiao Saltern on the coast of yellow sea of China. Evid. Based Complement. Altern. Med. 2011, 2011, 615032. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, S.B.; Sayyed, R.Z.; Trivedi, M.H.; Gobi, T.A. Phosphate solubilizing microbes: Sustainable approach for managing phosphorus deficiency in agricultural soils. Springerplus 2013, 2, 587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahemad, M.; Kibret, M. Mechanisms and applications of plant growth promoting rhizobacteria: Current perspective. J. King Saud Univ.-Sci. 2014, 26, 1–20. [Google Scholar] [CrossRef] [Green Version]
- Spaepen, S.; Vanderleyden, J.; Remans, R. Indole-3-acetic acid in microbial and microorganism-plant signaling. FEMS Microbiol. Rev. 2007, 31, 425–448. [Google Scholar] [CrossRef] [Green Version]
- Insuk, C.; Kuncharoen, N.; Cheeptham, N.; Tanasupawat, S.; Pathom-aree, W. Bryophytes harbor cultivable actinobacteria with plant growth promoting potential. Front. Microbiol. 2020, 11, 563047. [Google Scholar] [CrossRef]
- Kaewkla, O.; Suriyachadkun, C.; Milton, C.; Franco, M. Streptomyces adelaidensis sp. nov., an actinobacterium isolated from the root of Callitris preissii with potential for plant growth-promoting properties. Arch. Microbiol. 2021, 203, 3341–3352. [Google Scholar] [CrossRef]
- Subramaniam, G.; Thakur, V.; Saxena, R.K.; Vadlamudi, S.; Purohit, S.; Kumar, V.; Rathore, A.; Chitikineni, A.; Varshney, R.K. Complete genome sequence of sixteen plant growth promoting Streptomyces strains. Sci. Rep. 2020, 10, 10294. [Google Scholar] [CrossRef]
- Ouyang, J.; Shao, X.; Li, J. Indole-3-glycerol phosphate, a branchpoint of indole-3- acetic acid biosynthesis from the tryptophan biosynthetic pathway in Arabidopsis thaliana. Plant J. 2000, 24, 327–333. [Google Scholar] [CrossRef]
- Singh, S.P.; Gupta, R.; Gaur, R.; Srivastava, A.K. Antagonistic Actinomycetes Mediated Resistance in Solanum lycopersicon Mill. Against Rhizoctonia solani Kühn. Proc. Natl. Acad. Sci. India Sect. B-Biol. Sci. 2017, 87, 789–798. [Google Scholar] [CrossRef]
- Goudjal, Y.; Toumatia, O.; Sabaou, N.; Barakate, M.; Mathieu, F.; Zitouni, A. Endophytic actinomycetes from spontaneous plants of Algerian Sahara: Indole-3-acetic acid production and tomato plants growth promoting activity. World J. Microbiol. Biotechnol. 2013, 29, 1821–1829. [Google Scholar] [CrossRef] [Green Version]
- Andrews, S.C.; Robinson, A.K.; Rodríguez-Quiñones, F. Bacterial iron homeostasis. FEMS Microbiol. Rev. 2003, 27, 215–237. [Google Scholar] [CrossRef]
- Sandy, M.; Butler, A. Microbial Iron Acquisition: Marine and Terrestrial Siderophores. Chem. Rev. 2009, 109, 4580–4595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lau, C.K.Y.; Krewulak, K.D.; Vogel, H.J. Bacterial ferrous iron transport : The Feo system. FEMS Microbiol. Rev. 2015, 40, 273–298. [Google Scholar] [CrossRef] [PubMed]
- Dastager, S.G.; Damare, S. Marine actinobacteria showing phosphate-solubilizing efficiency in Chorao Island, Goa, India. Curr. Microbiol. 2013, 66, 421–427. [Google Scholar] [CrossRef] [PubMed]
- Santos-Beneit, F. The Pho regulon: A huge regulatory network in bacteria. Front. Microbiol. 2015, 6, 402. [Google Scholar] [CrossRef] [Green Version]
- Martín, J.F.; Liras, P. Molecular mechanisms of phosphate sensing, transport and signalling in streptomyces and related actinobacteria. Int. J. Mol. Sci. 2021, 22, 1129. [Google Scholar] [CrossRef]
- Rawat, P.; Das, S.; Shankhdhar, D.; Shankhdhar, S.C. Phosphate-solubilizing microorganisms: Mechanism and their role in phosphate solubilization and uptake. J. Soil Sci. Plant Nutr. 2021, 21, 49–68. [Google Scholar] [CrossRef]
- Luo, H.; Benner, R.; Long, R.A.; Hu, J. Subcellular localization of marine bacterial alkaline phosphatases. Proc. Natl. Acad. Sci. USA 2009, 106, 21219–21223. [Google Scholar] [CrossRef] [Green Version]
- Burkovski, A. Ammonium assimilation and nitrogen control in Corynebacterium glutamicum and its relatives: An example for new regulatory mechanisms in actinomycetes. FEMS Microbiol. Rev. 2003, 27, 617–628. [Google Scholar] [CrossRef] [Green Version]
- Tanveer, K.; Gilani, S.; Hussain, Z.; Ishaq, R.; Adeel, M.; Ilyas, N. Effect of salt stress on tomato plant and the role of calcium. J. Plant Nutr. 2019, 43, 28–35. [Google Scholar] [CrossRef]
- Barrs, H.; Weatherley, P. A re-examination of the relative turgidity technique for estimating water deficits in leaves. Aust. J. Biol. Sci. 1962, 15, 413–428. [Google Scholar] [CrossRef] [Green Version]
- Kempf, M.B.; Bremer, E. Uptake and synthesis of compatible solutes as microbial stress responses to high-osmolality environments. Arch. Microbiol. 1998, 170, 319–330. [Google Scholar] [CrossRef] [PubMed]
- León, M.J.; Hoffmann, T.; Sánchez-porro, C.; Heider, J.; Ventosa, A.; Bremer, E.; Oren, A. Compatible solute synthesis and import by the moderate halophile Spiribacter salinus: Physiology and genomics. Front. Microbiol. 2018, 9, 108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crowley, E. Compatible solute ectoine review: Protection mechanisms and production methods. J. Undergrad. Stud. Trent 2017, 5, 32–39. [Google Scholar]
- Czech, L.; Hermann, L.; Stöveken, N.; Richter, A.A.; Id, A.H.; Smits, S.H.J.; Heider, J.; Bremer, E. Role of the extremolytes ectoine and hydroxyectoine as stress protectants and nutrients: Genetics, phylogenomics, biochemistry, and structural analysis. Genes 2018, 9, 177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bursy, J.; Kuhlmann, A.U.; Pittelkow, M.; Hartmann, H.; Jebbar, M.; Pierik, A.J.; Bremer, E. Synthesis and uptake of the compatible solutes ectoine and 5-hydroxyectoine by Streptomyces coelicolor A3 (2) in response to salt and heat stresses. Appl. Environ. Microbiol. 2008, 74, 7286–7296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wargo, M.J. Homeostasis and catabolism of choline and glycine betaine: Lessons from Pseudomonas aeruginosa. Appl. Environ. Microbiol. 2013, 79, 2112–2120. [Google Scholar] [CrossRef] [Green Version]
- Kappes, R.M.; Kempf, B.; Bremer, E. Three transport systems for the osmoprotectant glycine betaine pperate in Bacillus subtilis: Characterization of OpuD. J. Bacteriol. 1996, 178, 5071–5079. [Google Scholar] [CrossRef] [Green Version]
- Lamark, T.; Kaasen, I.; Eshoo, M.W.; Falkenberg, P.; Mcdougall, J.; Strom, A.R. DNA sequence and analysis of the bef genes encoding the osmoregulatory choline-glycine betaine pathway of Escherichia coli B. Mol. Microbiol. 1991, 5, 1049–1064. [Google Scholar] [CrossRef]
- Ansari, M.; Shekari, F.; Mohammadi, M.H.; Juhos, K.; Végvári, G.; Biró, B. Salt-tolerant plant growth-promoting bacteria enhanced salinity tolerance of salt-tolerant alfalfa (Medicago sativa L.) cultivars at high salinity. Acta Physiol. Plant. 2019, 41, 195. [Google Scholar] [CrossRef] [Green Version]
- Avonce, N.; Mendoza-vargas, A.; Morett, E.; Iturriaga, G. Insights on the evolution of trehalose biosynthesis. BMC Evol. Biol. 2006, 6, 109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ben Rejeb, K.; Abdelly, C.; Savouré, A. How reactive oxygen species and proline face stress together. Plant Physiol. Biochem. 2014, 80, 278–284. [Google Scholar] [CrossRef] [PubMed]
- Jamali, S.S.; Borzouei, A.; Aghamirzaei, M.; Khosronejad, H.R.; Fathi, M. Cell membrane stability and biochemical response of seven wheat cultivars under salinity stress. Rev. Bras. Bot. 2015, 38, 63–69. [Google Scholar] [CrossRef]
- Kaushal, M.; Wani, S.P. Plant-growth-promoting rhizobacteria: Drought stress alleviators to ameliorate crop production in drylands. Ann. Microbiol. 2016, 66, 35–42. [Google Scholar] [CrossRef]
- Fichman, Y.; Gerdes, S.Y.; Kovács, H.; Szabados, L.; Zilberstein, A.; Csonka, L.N. Evolution of proline biosynthesis: Enzymology, bioinformatics, genetics, and transcriptional regulation. Biol. Rev. 2015, 90, 1065–1099. [Google Scholar] [CrossRef]
- Liang, Q.J.; Wu, X.J.; Yang, P.; Kong, J.R.; Wei, W.; Qiao, X.; Liu, Y.; Wang, W. The role of delta-1-pyrroline-5-carboxylate dehydrogenase (P5CDh) in the Pacific white shrimp (Litopenaeus vannamei) during biotic and abiotic stress. Aquat. Toxicol. 2019, 208, 1–11. [Google Scholar] [CrossRef]
- Rojas-tapias, D.; Moreno-galván, A.; Pardo-díaz, S.; Obando, M.; Rivera, D.; Bonilla, R. Effect of inoculation with plant growth-promoting bacteria (PGPB) on amelioration of saline stress in maize (Zea mays). Appl. Soil Ecol. 2012, 61, 264–272. [Google Scholar] [CrossRef]
- Islam, F.; Yasmeen, T.; Arif, M.S.; Ali, S. Plant growth promoting bacteria confer salt tolerance in Vigna radiata by up-regulating antioxidant defense and biological soil fertility. Plant Growth Regul. 2016, 80, 23–36. [Google Scholar] [CrossRef]
- Curá, J.A.; Franz, D.R.; Filosofía, J.E.; Balestrasse, K.B.; Burgueño, L.E. Inoculation with Azospirillum sp. and Herbaspirillum sp. bacteria increases the tolerance of maize to drought stress. Microorganisms 2017, 5, 41. [Google Scholar] [CrossRef] [Green Version]
- Abbasi, S.; Sadeghi, A.; Safaie, N. Streptomyces alleviate drought stress in tomato plants and modulate the expression of transcription factors ERF1 and WRKY70 genes. Sci. Hortic. 2020, 265, 109206. [Google Scholar] [CrossRef]
- Masmoudi, F.; Tounsi, S.; Dunlap, C.A.; Trigui, M. Endophytic halotolerant Bacillus velezensis FMH2 alleviates salt stress on tomato plants by improving plant growth and altering physiological and antioxidant responses. Plant Physiol. Biochem. 2021, 165, 217–227. [Google Scholar] [CrossRef] [PubMed]
- Cha, M.; Bae, Y.; Kim, K.; Park, B.; Kim, I.; Cha, M.; Bae, Y.; Kim, K. Characterization of two alkyl hydroperoxide reductase C homologs alkyl hydroperoxide reductase C_H1 and alkyl hydroperoxide reductase C_H2 in Bacillus subtilis. World J. Biol. Chem. 2015, 6, 249–265. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Chung, C.; Ma, T.; Wong, H. Roles of alkyl hydroperoxide reductase subunit C (AhpC) in viable but nonculturable Vibrio parahaemolyticus. Appl. Environ. Microbiol. 2013, 79, 3734–3743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chattopadhyay, M.K.; Chen, W.; Tabor, H. Escherichia coli glutathionylspermidine synthetase/amidase: Phylogeny and effect on regulation of gene expression. FEMS Microbiol. Lett. 2013, 338, 132–140. [Google Scholar] [CrossRef] [Green Version]
- Kwon, D.S.; Lin, C.; Chen, S.; Coward, J.K.; Walsh, C.T.; Bollinger, J.M. Dissection of glutathionylspermidine synthetase/amidase from Escherichia coli into autonomously folding and functional synthetase and amidase domains. J. Biol. Chem. 1997, 272, 2429–2436. [Google Scholar] [CrossRef] [Green Version]
- Machado, R.; Serralheiro, R. Soil salinity: Effect on vegetable crop growth. management practices to prevent and mitigate soil salinization. Horticulturae 2017, 3, 30. [Google Scholar] [CrossRef]
- Al-aghabary, K.; Zhu, Z.; Shi, Q. Influence of silicon supply on chlorophyll content, chlorophyll fluorescence, and antioxidative enzyme activities in tomato plants under salt stress antioxidative enzyme activities in tomato. J. Plant Nutr. 2007, 27, 2101–2115. [Google Scholar] [CrossRef]
- Da Silva, A.A.R.; de Lima, G.S.; de Azevedo, C.A.V.; de S.A. Veloso, L.L.; Gheyi, H.R.; dos A. Soares, L.A. Salt stress and exogenous application of hydrogen peroxide on photosynthetic parameters of soursop. Rev. Bras. Eng. Agrícola Ambient. 2019, 23, 257–263. [Google Scholar] [CrossRef] [Green Version]
- Orozco-mosqueda, M.C.; Duan, J.; Dibernardo, M.; Zetter, E.; Campos-garcía, J.; Glick, B.R.; Santoyo, G. The production of ACC deaminase and trehalose by the plant growth promoting bacterium Pseudomonas sp. UW4 synergistically protect tomato plants against salt stress. Front. Microbiol. 2019, 10, 1392. [Google Scholar] [CrossRef] [Green Version]
- Darby, C.; Cosma, C.L.; Thomas, J.H.; Manoil, C. Lethal paralysis of Caenorhabditis elegans by Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 1999, 96, 15202–15207. [Google Scholar] [CrossRef] [Green Version]
- Harrington, A.J.; Hamamichi, S.; Caldwell, G.A.; Caldwell, K.A. C. elegans as a model organism to investigate molecular pathways involved with Parkinson’s disease. Dev. Dyn. 2010, 239, 1282–1295. [Google Scholar] [CrossRef] [PubMed]
- Park, H.H.; Jung, Y.; Lee, S.V. Molecules and Cells Survival assays using Caenorhabditis elegans. Mol. Cells 2017, 40, 90–99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maglioni, S.; Ventura, N. C. elegans as a model organism for human mitochondrial associated disorders. Mitochondrion 2016, 30, 117–125. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Li, F.; Zhou, T.; Wang, G.; Li, Z. Caenorhabditis elegans as a Useful Model for Studying Aging Mutations. Front. Endocrinol. 2020, 11, 554994. [Google Scholar] [CrossRef] [PubMed]
- Marsh, E.K.; May, R.C. Caenorhabditis elegans, A model organism for investigating immunity. Appl. Environ. Microbiol. 2012, 78, 2075–2081. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Darvesh, A.S.; Barnett, R.E.; Fitsanakis, V.A. Caenorhabditis elegans: An elegant model organism for evaluating the neuroprotective and neurotherapeutic potential of nutraceuticals. In Nutraceuticals; Gupta, R.C., Lall, R., Srivastava, A., Eds.; Academic Press: London, UK, 2021; pp. 411–430. [Google Scholar] [CrossRef]
- De Sousa Figueiredo, M.B.; Pradel, E.; George, F.; Mahieux, S.; Houcke, I.; Pottier, M.; Fradin, C.; Neut, C.; Daniel, C.; Bongiovanni, A.; et al. Adherent-Invasive and Non-Invasive Escherichia coli Isolates Differ in Their Effects on Caenorhabditis elegans’ Lifespan. Microorganisms 2021, 9, 1823. [Google Scholar] [CrossRef]
- Sánchez-Diener, I.; Zamorano, L.; López-Causapé, C.; Cabot, G.; Mulet, X.; Peña, C.; del Campo, R.; Cantón, R.; Doménech-Sánchez, A.; Martínez-Martínez, L.; et al. Interplay among Resistance Profiles, High-Risk Clones, and Virulence in the Caenorhabditis elegans Pseudomonas aeruginosa Infection Model. Antimicrob. Agents Chemother. 2017, 61, e01586-17. [Google Scholar] [CrossRef] [Green Version]
- Keswani, C.; Prakash, O.; Bharti, N.; Vílchez, J.I.; Sansinenea, E.; Lally, R.D.; Borriss, R.; Singh, S.P.; Gupta, V.K.; Fraceto, L.F.; et al. Re-addressing the biosafety issues of plant growth promoting rhizobacteria. Sci. Total Environ. 2019, 690, 841–852. [Google Scholar] [CrossRef]





| NaCl Concentration (mM) | IAA Production (µg mL−1) | Siderophore Production (µmol mL−1) | Phosphate Solubilization | ||
|---|---|---|---|---|---|
| Hydroxamate | Catecholate | P Released in PVK Broth (µg mL−1) | pH | ||
| 0 | 37.50 a ± 1.61 | 46.67 a ± 17.56 | 2.98 a ± 2.90 | 71.62 a ± 3.02 | 5.1 ± 0.07 |
| 150 | 11.75 b ± 0.35 | 173.33 b ± 31.66 | 48.25 b ± 17.87 | 67.98 ab ± 1.41 | 5.4 ± 0.03 |
| 300 | 8.36 b ± 0.32 | 189.17 b ± 31.75 | 21.58 c ± 2.73 | 67.09 b ± 1.79 | 5.4 ± 0.04 |
| 450 | 10.55 b ± 5.39 | 170.83 b ± 3.82 | 20.70 c ± 1.69 | 62.33c ± 2.13 | 5.4 ± 0.05 |
| PGP Traits | Protein Coding Sequences Conferring PGP Traits |
|---|---|
| Amino Acids and Derivatives | Proline synthesis:
|
Tryptophan synthesis:
| |
| |
| Iron acquisition and metabolism | Ferrous iron transporter EfeUOB, low-pH-induced:
|
| Phosphorus metabolism | High affinity phosphate transporter and control of PHO regulon:
|
| N2 metabolism | Ammonia assimilation:
|
| Trehalose metabolism | Trehalose biosynthesis:
|
| Potassium metabolism | Potassium homeostasis:
|
| Osmotic stress response | Osmoregulation: Glycerol uptake facilitator proteinEctoine biosynthesis and regulation: L-ectoine synthase (EC 4.2.1.-)Choline and Betaine Uptake and Betaine Biosynthesis:
|
| Oxidative stress response | Alkyl hydroperoxide reductase subunit C-like proteinGlutathionylspermidine and Trypanothione:
|
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rangseekaew, P.; Barros-Rodríguez, A.; Pathom-aree, W.; Manzanera, M. Plant Beneficial Deep-Sea Actinobacterium, Dermacoccus abyssi MT1.1T Promote Growth of Tomato (Solanum lycopersicum) under Salinity Stress. Biology 2022, 11, 191. https://doi.org/10.3390/biology11020191
Rangseekaew P, Barros-Rodríguez A, Pathom-aree W, Manzanera M. Plant Beneficial Deep-Sea Actinobacterium, Dermacoccus abyssi MT1.1T Promote Growth of Tomato (Solanum lycopersicum) under Salinity Stress. Biology. 2022; 11(2):191. https://doi.org/10.3390/biology11020191
Chicago/Turabian StyleRangseekaew, Pharada, Adoración Barros-Rodríguez, Wasu Pathom-aree, and Maximino Manzanera. 2022. "Plant Beneficial Deep-Sea Actinobacterium, Dermacoccus abyssi MT1.1T Promote Growth of Tomato (Solanum lycopersicum) under Salinity Stress" Biology 11, no. 2: 191. https://doi.org/10.3390/biology11020191
APA StyleRangseekaew, P., Barros-Rodríguez, A., Pathom-aree, W., & Manzanera, M. (2022). Plant Beneficial Deep-Sea Actinobacterium, Dermacoccus abyssi MT1.1T Promote Growth of Tomato (Solanum lycopersicum) under Salinity Stress. Biology, 11(2), 191. https://doi.org/10.3390/biology11020191