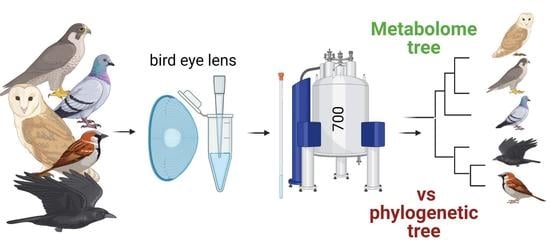
The Application of Quantitative Metabolomics for the Taxonomic Differentiation of Birds
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Lens Sample Collection and Species Description
2.3. Sample Preparation
2.4. NMR Measurements
2.5. LC–MS Measurements
2.6. Metabolomic Data Analysis
2.7. Phylogenetic Tree Reconstructions for Birds from the Literature
3. Results
3.1. The Identification and Quantification of Metabolites
3.2. General Overview of Bird Lens Metabolomes
3.3. Principal Component Analysis (PCA)
3.4. Hierarchical Clustering Analysis (HCA) and Heatmaps
3.5. Genomics- and Transcriptomics-Based Schematic Phylogenetic Tree Construction from the Literature
4. Discussion
- Conspecific samples are positioned together and form clusters separated from other species (Figure 3, Supplementary Figure S3 and S4).
- All four Passerides (C. coccothraustes, E. godlewskii, P. domesticus, P. major) are positioned together in a separate larger cluster.
- All four Corvides (P. pica, C. corax, C. cornix, C. frugilegus) are positioned together in a single cluster.
- The Passerides and the Corvides samples are positioned in two connected branches, forming a larger cluster (Passeriformes).
- C. livia and P. cristatus are distant from other clusters.
- A node between C. livia and P. cristatus from the Columbea clade in the Jarvis ED et al. tree [24] does not exist in any HCA dendrogram (dashed green lilac arrow).
- F. atra in the HCA dendrogram is positioned close to the Corvides infraorder, most likely indicating the influence of lifestyle on the metabolomic composition of the F. atra eye lens (dark violet arrow); no node between F. atra and C. livia (the Basal landbirds clade from Kuhl H et al. tree [27]) was found in any HCA dendrogram.
- Although C. frugilegus and P. pica are well clustered with the Corvides, they have rather incorrect phylogenetic distances within the Corvides. C. frugilegus should be closer to the other species from the Corvus genus (C. corax and C. cornix), and P. pica should be the sister taxon to all Corvus. The samples of P. pica, C. corax and C. cornix often mix together, without the formation of separate clusters for each species (dashed magenta, mint and aquamarine arrows).
- Similarly, the incorrect positioning is observed for P. major; it should be more distant from the other species of the Passerida parvorder (light green arrow).
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Livezey, B.C.; Zusi, R.L. Higher-order phylogeny of modern birds (Theropoda, Aves: Neornithes) based on comparative anatomy. II. Analysis and discussion. Zool. J. Linn. Soc. 2007, 149, 1–95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hennig, W. Phylogenetic Systematics; University of Illinois Press: Champaign, IL, USA, 1999; ISBN 978-0-252-06814-0. [Google Scholar]
- Brown, T.A. Molecular Phylogenetics. In Genomes 3; Brown, T.A., Ed.; Garland Science Publishing: New York, NY, USA, 2007. [Google Scholar]
- Ningthoujam, S.S.; Talukdar, A.D.; Sarker, S.D.; Nahar, L.; Choudhury, M.D. Prediction of Medicinal Properties Using Mathematical Models and Computation, and Selection of Plant Materials. In Computational Phytochemistry; Sarker, S.D., Nahar, L., Eds.; Elsevier: Amsterdam, The Netherlands, 2018; pp. 43–73. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Suleski, M.; Hedges, S.B. TimeTree: A Resource for Timelines, Timetrees, and Divergence Times. Mol. Biol. Evol. 2017, 34, 1812–1819. [Google Scholar] [CrossRef] [PubMed]
- Dayrat, B. Towards integrative taxonomy. Biol. J. Linn. Soc. 2005, 85, 407–415. [Google Scholar] [CrossRef]
- Padial, J.M.; Miralles, A.; De La Riva, I.; Vences, M. The integrative future of taxonomy. Front. Zool. 2010, 7, 16. [Google Scholar] [CrossRef]
- Daglio, L.G.; Dawson, M.N. Integrative taxonomy: Ghosts of past, present and future. J. Mar. Biol. Assoc. 2019, 99, 1237–1246. [Google Scholar] [CrossRef]
- Ccana-Ccapatinta, G.V.; Feitas, J.A.; Monge, M.; Ferreira, P.d.L.; Semir, J.; Groppo, M.; Da Costa, F.B. Metabolomics and chemophenetics support the new taxonomy circumscription of two South America genera (Barnadesioideae, Asteraceae). Phytochem. Lett. 2020, 40, 89–95. [Google Scholar] [CrossRef]
- Cheng, Z.; Shu, H.; Zhang, S.; Luo, B.; Gu, R.; Zhang, R.; Ji, Y.; Li, F.; Long, C. From Folk Taxonomy to Species Confirmation of Acorus (Acoraceae): Evidences Based on Phylogenetic and Metabolomic Analyses. Front. Plant Sci. 2020, 11, 965. [Google Scholar] [CrossRef]
- Chervin, J.; Talou, T.; Audonnet, M.; Dumas, B.; Camborde, L.; Esquerré-Tugayé, M.-T.; Roux, C.; Cabanac, G.; Marti, G. Deciphering the phylogeny of violets based on multiplexed genetic and metabolomic approaches. Phytochemistry 2019, 163, 99–110. [Google Scholar] [CrossRef] [Green Version]
- Farag, M.A.; Weigend, M.; Luebert, F.; Brokamp, G.; Wessjohann, L.A. Phytochemical, phylogenetic, and anti-inflammatory evaluation of 43 Urtica accessions (stinging nettle) based on UPLC–Q-TOF-MS metabolomic profiles. Phytochemistry 2013, 96, 170–183. [Google Scholar] [CrossRef]
- Martucci, M.E.P.; Loeuille, B.; Pirani, J.R.; Gobbo-Neto, L. Comprehensive untargeted metabolomics of Lychnnophorinae subtribe (Asteraceae: Vernonieae) in a phylogenetic context. PLoS ONE 2018, 13, e0190104. [Google Scholar] [CrossRef] [Green Version]
- Dos Santos, V.S.; Macedo, F.A.; Vale, J.S.D.; da Silva, D.; Carollo, C.A. Metabolomics as a tool for understanding the evolution of Tabebuia sensu lato. Metabolomics 2017, 13, 72. [Google Scholar] [CrossRef]
- Kim, H.K.; Saifullah; Khan, S.; Wilson, E.G.; Kricun, S.D.P.; Meissner, A.; Goraler, S.; Deelder, A.M.; Choi, Y.H.; Verpoorte, R. Metabolic classification of South American Ilex species by NMR-based metabolomics. Phytochemistry 2010, 71, 773–784. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.; Peever, T.L.; Park, J.-J.; Park, C.-M.; Gang, D.R.; Xian, M.; Davidson, J.A.; Infantino, A.; Kaiser, W.J.; Chen, W. Use of metabolomics for the chemotaxonomy of legume-associated Ascochyta and allied genera. Sci. Rep. 2016, 6, 20192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Y.-T.; Skelton, J.; Hulcr, J. Lipids and small metabolites provisioned by ambrosia fungi to symbiotic beetles are phylogeny-dependent, not convergent. ISME J. 2020, 14, 1089–1099. [Google Scholar] [CrossRef] [PubMed]
- Gregor, R.; Probst, M.; Eyal, S.; Aksenov, A.; Sasson, G.; Horovitz, I.; Dorrestein, P.C.; Meijler, M.M.; Mizrahi, I. Mammalian gut metabolomes mirror microbiome composition and host phylogeny. ISME J. 2021, 16, 1262–1274. [Google Scholar] [CrossRef]
- Han, S.; Van Treuren, W.; Fischer, C.R.; Merrill, B.D.; DeFelice, B.C.; Sanchez, J.M.; Higginbottom, S.K.; Guthrie, L.; Fall, L.A.; Dodd, D.; et al. A metabolomics pipeline for the mechanistic interrogation of the gut microbiome. Nature 2021, 595, 415–420. [Google Scholar] [CrossRef]
- Sibley, C.G.; Ahlquist, J.E. Phylogeny and Classification of the Birds: A Study in Molecular Evolution; Yale University Press: London, UK, 1990; ISBN 978-0-300-23785-6. [Google Scholar]
- Ericson, P.G.; Anderson, C.L.; Britton, T.; Elzanowski, A.; Johansson, U.S.; Källersjö, M.; Ohlson, J.I.; Parsons, T.J.; Zuccon, D.; Mayr, G. Diversification of Neoaves: Integration of molecular sequence data and fossils. Biol. Lett. 2006, 2, 543–547. [Google Scholar] [CrossRef]
- Hackett, S.J.; Kimball, R.T.; Reddy, S.; Bowie, R.C.K.; Braun, E.L.; Braun, M.J.; Chojnowski, J.L.; Cox, W.A.; Han, K.-L.; Harshman, J.; et al. A Phylogenomic Study of Birds Reveals Their Evolutionary History. Science 2008, 320, 1763–1768. [Google Scholar] [CrossRef]
- Jetz, W.; Thomas, G.; Joy, J.B.; Hartmann, K.; Mooers, A.O. The global diversity of birds in space and time. Nature 2012, 491, 444–448. [Google Scholar] [CrossRef]
- Jarvis, E.D.; Mirarab, S.; Aberer, A.J.; Li, B.; Houde, P.; Li, C.; Ho, S.Y.W.; Faircloth, B.C.; Nabholz, B.; Howard, J.T.; et al. Whole-genome analyses resolve early branches in the tree of life of modern birds. Science 2014, 346, 1320–1331. [Google Scholar] [CrossRef] [Green Version]
- Prum, R.O.; Berv, J.S.; Dornburg, A.; Field, D.J.; Townsend, J.P.; Lemmon, E.M.; Lemmon, A.R. A comprehensive phylogeny of birds (Aves) using targeted next-generation DNA sequencing. Nature 2015, 526, 569–573. [Google Scholar] [CrossRef] [PubMed]
- Kimball, R.T.; Oliveros, C.H.; Wang, N.; White, N.D.; Barker, F.K.; Field, D.J.; Ksepka, D.T.; Chesser, R.T.; Moyle, R.G.; Braun, M.J.; et al. A Phylogenomic Supertree of Birds. Diversity 2019, 11, 109. [Google Scholar] [CrossRef] [Green Version]
- Kuhl, H.; Frankl-Vilches, C.; Bakker, A.; Mayr, G.; Nikolaus, G.; Boerno, S.T.; Klages, S.; Timmermann, B.; Gahr, M. An Unbiased Molecular Approach Using 3′-UTRs Resolves the Avian Family-Level Tree of Life. Mol. Biol. Evol. 2020, 38, 108–127. [Google Scholar] [CrossRef] [PubMed]
- Colbert, E.H.; Edwin, H. Evolution of the Vertebrates: A History of the Backboned Animals through Time, 3rd ed.; Wiley: Hoboken, NJ, USA, 1980; ISBN 978-0471049661. [Google Scholar]
- Suh, A. The phylogenomic forest of bird trees contains a hard polytomy at the root of Neoaves. Zool. Scr. 2016, 45, 50–62. [Google Scholar] [CrossRef]
- Thomas, G.H. Evolution: An Avian Explosion. Nature 2015, 526, 516–517. [Google Scholar] [CrossRef] [PubMed]
- Braun, E.; Kimball, R. Data Types and the Phylogeny of Neoaves. Birds 2021, 2, 1–22. [Google Scholar] [CrossRef]
- Forrester, J.V.; Dick, A.D.; McMenamin, P.G.; Roberts, F.; Eric Pearlman, B.S. The Eye: Basic Sciences in Practice; ClinicalKey 2012; Elsevier Health Sciences: Amsterdam, The Netherlands, 2015; ISBN 9780702055546. [Google Scholar]
- Yanshole, V.V.; Snytnikova, O.A.; Kiryutin, A.S.; Yanshole, L.V.; Sagdeev, R.Z.; Tsentalovich, Y.P. Metabolomics of the rat lens: A combined LC-MS and NMR study. Exp. Eye Res. 2014, 125, 71–78. [Google Scholar] [CrossRef]
- Yanshole, V.V.; Yanshole, L.V.; Snytnikova, O.A.; Tsentalovich, Y.P. Quantitative metabolomic analysis of changes in the lens and aqueous humor under development of age-related nuclear cataract. Metabolomics 2019, 15, 29. [Google Scholar] [CrossRef]
- Tsentalovich, Y.P.; Verkhovod, T.D.; Yanshole, V.; Kiryutin, A.; Yanshole, L.; Fursova, A.Z.; Stepakov, D.A.; Novoselov, V.P.; Sagdeev, R.Z. Metabolomic composition of normal aged and cataractous human lenses. Exp. Eye Res. 2015, 134, 15–23. [Google Scholar] [CrossRef]
- Tsentalovich, Y.P.; Yanshole, V.V.; Zelentsova, E.A.; Melnikov, A.D.; Sagdeev, R.Z. Seasonal Variations and Interspecific Differences in Metabolomes of Freshwater Fish Tissues: Quantitative Metabolomic Profiles of Lenses and Gills. Metabolites 2019, 9, 264. [Google Scholar] [CrossRef] [Green Version]
- Edison, A.S.; Hall, R.D.; Junot, C.; Karp, P.D.; Kurland, I.J.; Mistrik, R.; Reed, L.K.; Saito, K.; Salek, R.M.; Steinbeck, C.; et al. The Time Is Right to Focus on Model Organism Metabolomes. Metabolites 2016, 6, 8. [Google Scholar] [CrossRef] [Green Version]
- Gill, F.; Donsker, D.; Rasmussen, P. (Eds.) IOC World Bird List 12.1; 2022. [Google Scholar] [CrossRef]
- Tsentalovich, Y.P.; Zelentsova, E.A.; Yanshole, L.V.; Yanshole, V.V.; Odud, I.M. Most abundant metabolites in tissues of freshwater fish pike-perch (Sander lucioperca). Sci. Rep. 2020, 10, 17128. [Google Scholar] [CrossRef] [PubMed]
- Yanshole, V.V.; Yanshole, L.V.; Zelentsova, E.A.; Tsentalovich, Y.P. Ovothiol A is the Main Antioxidant in Fish Lens. Metabolites 2019, 9, 95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pang, Z.; Chong, J.; Zhou, G.; de Lima Morais, D.A.; Chang, L.; Barrette, M.; Gauthier, C.; Jacques, P.-É.; Li, S.; Xia, J. MetaboAnalyst 5.0: Narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021, 49, W388–W396. [Google Scholar] [CrossRef] [PubMed]
- Oliveros, C.H.; Field, D.J.; Ksepka, D.T.; Barker, F.K.; Aleixo, A.; Andersen, M.J.; Alström, P.; Benz, B.W.; Braun, E.L.; Braun, M.J.; et al. Earth history and the passerine superradiation. Proc. Natl. Acad. Sci. USA 2019, 116, 7916–7925. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ripps, H.; Shen, W. Review: Taurine: A “Very Essential” Amino Acid. Mol. Vis. 2012, 18, 2673. [Google Scholar]
- Jiang, Z.; Chung, S.K.; Zhou, C.; Cammarata, P.R. Overexpression of Na+-dependent myo-inositol transporter gene in mouse lens led to congenital cataract. Investig. Ophthalmol. Vis. Sci. 2000, 41, 1467–1472. [Google Scholar]
- Jones, M.P.; Pierce, K.E.; Ward, D. Avian Vision: A Review of Form and Function with Special Consideration to Birds of Prey. J. Exot. Pet Med. 2007, 16, 69–87. [Google Scholar] [CrossRef]
- Wehrens, R.; Salek, R. Metabolomics: Practical Guide to Design and Analysis; CRC Press: Boca Raton, FL, USA, 2019; ISBN 1498725279. [Google Scholar]
- Verpoorte, R.; Choi, Y.H.; Mustafa, N.R.; Kim, H.K. Metabolomics: Back to Basics. Phytochem. Rev. 2008, 7, 525–537. [Google Scholar] [CrossRef]
- Serebryany, E.; Chowdhury, S.; Woods, C.N.; Thorn, D.C.; Watson, N.E.; McClelland, A.A.; Klevit, R.E.; Shakhnovich, E.I. A native chemical chaperone in the human eye lens. eLife 2022, 11, e76923. [Google Scholar] [CrossRef]





| Species * | Date and Place of Catching | Typical Lens or Sample Weight, mg | N |
|---|---|---|---|
| Black kite (Milvus migrans) | July 2019, Tyva Republic | 90–150 | 5 individuals |
| Eurasian magpie (Pica pica) | December 2018–January 2019, Altay Republic | 90–125 | 4 individuals |
| Northern raven (Corvus corax) | December 2018, Altay Republic; July 2019, Tyva Republic; December 2019, Novosibirsk Region | 130–240 | 5 individuals |
| Eurasian coot (Fulica atra) | April 2019, Novosibirsk Region; May 2019, Tyva Republic | 50–90 | 4 individuals |
| Godlewski’s bunting (Emberiza godlewskii) | January 2019, Altay Republic | 60–70 per sample; 20–25 per lens | 18 individuals, 6 samples |
| Great crested grebe (Podiceps cristatus) | May 2019, Tyva Republic | 70–80 | 5 individuals |
| Great tit (Parus major) | December 2018, Altay Republic | 40–90 | 12 individuals, 6 samples |
| Hawfinch (Coccothraustes coccothraustes) | December 2018, Altay Republic | 70–110 per sample; 30–40 per lens | 6 individuals, 3 samples |
| Hooded crow (Corvus cornix) | January 2019, Altay Republic; April 2019, Novosibirsk Region | 110–200 | 3 individuals |
| House sparrow (Passer domesticus) | November 2018, Novosibirsk Region | 35–65 per sample; 20–30 per lens | 14 individuals, 7 samples |
| Rock dove (Columba livia) | September 2017, Novosibirsk Region | 25–55 | 12 individuals |
| Rook (Corvus frugilegus) | April 2019, Novosibirsk Region | 60–105 | 5 individuals |
| Short-eared owl (Asio flammeus) | May 2019, CRWA, Novosibirsk | 156 | 1 individual |
| Ural owl (Strix uralensis) | May 2019, CRWA, Novosibirsk | 164 | 1 individual |
| Species | Black Kite | Common Magpie | Common Raven | Eurasian Coot | Godlewski’s Bunting | Great Crested Grebe | Great Tit | Hawfinch | Hooded Crow | House Sparrow | Rock Dove | Rook | Short-Eared Owl 1 | Ural Owl 1 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Species Latin | M. migrans | P. pica | C. corax | F. atra | E. godlewskii | P. cristatus | P. major | C. coccothraustes | C. cornix | P. domesticus | C. livia | C. frugilegus | A. flammeus | S. uralensis |
| 2,3-Butanediol | 91 ± 48 | 56 ± 50 | 31 ± 35 | 21 ± 17 | 19 ± 9 | 0 | 11 ± 7 | 11 ± 9 | 10 ± 4 | 130 ± 30 | 78 ± 34 | 0 | 0 | 0 |
| 2-Ketoisovalerate | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 21 | 9 |
| 2-OH-3-Me-but 2,3 | 18 ± 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 31 | 18 |
| 2-OH-but | 28 ± 12 | 18 ± 6 | 16 ± 5 | 14 ± 4 | 0 | 0 | 0 | 0 | 16 ± 3 | 0 | 0 | 0 | 40 | 13 |
| 3-Me-His 2 | 0 | 0 | 0 | 0 | 0 | 100 ± 30 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 3-OH-but | 96 ± 54 | 300 ± 130 | 210 ± 100 | 170 ± 40 | 250 ± 50 | 170 ± 50 | 370 ± 140 | 440 ± 40 | 340 ± 280 | 240 ± 60 | 91 ± 62 | 210 ± 180 | 480 | 480 |
| 3-OH-isovalerate | 0 | 34 ± 8 | 22 ± 7 | 66 ± 15 | 48 ± 10 | 51 ± 12 | 51 ± 24 | 10 ± 4 | 24 ± 7 | 44 ± 22 | 190 ± 60 | 47 ± 12 | 45 | 21 |
| Acetate | 4300 ± 1000 | 4300 ± 800 | 3000 ± 800 | 6000 ± 900 | 8100 ± 1200 | 5100 ± 600 | 9700 ± 2700 | 6200 ± 1800 | 3100 ± 1400 | 10,000 ± 3000 | 89 ± 27 | 4900 ± 1000 | 7200 | 3100 |
| Acetylcarnitine | 60 ± 22 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 19 | 22 |
| ADP | 560 ± 80 | 430 ± 40 | 380 ± 80 | 630 ± 120 | 960 ± 170 | 720 ± 50 | 1000 ± 200 | 740 ± 90 | 300 ± 100 | 1100 ± 200 | 470 ± 110 | 560 ± 150 | 290 | 470 |
| Alanine | 1700 ± 400 | 3000 ± 300 | 3200 ± 400 | 2100 ± 300 | 3600 ± 600 | 2400 ± 1100 | 6000 ± 800 | 5700 ± 100 | 2600 ± 1200 | 4700 ± 700 | 1700 ± 600 | 2900 ± 400 | 4900 | 1200 |
| alpha-Aminobut | 63 ± 11 | 56 ± 14 | 90 ± 26 | 130 ± 110 | 72 ± 20 | 220 ± 90 | 97 ± 45 | 91 ± 6 | 84 ± 12 | 120 ± 20 | 20 ± 6 | 53 ± 11 | 190 | 90 |
| alpha-OH-isobut 2 | 0 | 11 ± 4 | 11 ± 8 | 14 ± 14 | 0 | 0 | 0 | 0 | 12 ± 7 | 0 | 0 | 0 | 60 | 61 |
| AMP | 60 ± 37 | 28 ± 9 | 11 ± 9 | 31 ± 15 | 12 ± 8 | 51 ± 7 | 60 ± 36 | 10 ± 5 | 10 ± 2 | 280 ± 210 | 48 ± 47 | 39 ± 21 | 0 | 230 |
| Anserine | 120 ± 50 | 220 ± 160 | 290 ± 90 | 2400 ± 500 | 260 ± 20 | 0 | 73 ± 15 | 200 ± 30 | 240 ± 40 | 150 ± 60 | 0 | 750 ± 260 | 28 | 130 |
| Ascorbate | 390 ± 90 | 210 ± 40 | 230 ± 50 | 120 ± 20 | 440 ± 50 | 490 ± 70 | 190 ± 50 | 260 ± 10 | 160 ± 40 | 550 ± 70 | 200 ± 40 | 180 ± 20 | 480 | 370 |
| Asparagine | 170 ± 30 | 130 ± 70 | 120 ± 30 | 200 ± 50 | 0 | 37 ± 17 | 0 | 0 | 150 ± 30 | 0 | 0 | 54 ± 26 | 260 | 110 |
| Aspartate | 0 | 140 ± 50 | 210 ± 30 | 180 ± 40 | 130 ± 20 | 220 ± 70 | 160 ± 10 | 93 ± 14 | 220 ± 100 | 130 ± 30 | 44 ± 21 | 240 ± 70 | 100 | 89 |
| ATP | 2200 ± 300 | 3300 ± 200 | 3200 ± 300 | 3300 ± 200 | 2400 ± 100 | 3100 ± 300 | 4500 ± 1100 | 3100 ± 600 | 2700 ± 500 | 3200 ± 600 | 3800 ± 500 | 3100 ± 600 | 2900 | 1600 |
| Betaine | 590 ± 130 | 380 ± 60 | 900 ± 280 | 210 ± 70 | 1000 ± 200 | 230 ± 100 | 640 ± 130 | 720 ± 80 | 470 ± 60 | 800 ± 160 | 0 | 650 ± 230 | 230 | 110 |
| Carnitine | 43 ± 10 | 39 ± 7 | 100 ± 20 | 51 ± 12 | 0 | 52 ± 13 | 0 | 0 | 40 ± 4 | 0 | 27 ± 4 | 57 ± 7 | 140 | 110 |
| Carnosine | 0 | 60 ± 32 | 41 ± 32 | 550 ± 760 | 0 | 0 | 0 | 0 | 60 ± 13 | 0 | 0 | 98 ± 77 | 120 | 27 |
| Choline | 54 ± 8 | 11 ± 4 | 19 ± 14 | 78 ± 26 | 13 ± 2 | 85 ± 15 | 20 ± 8 | 32 ± 3 | 8.7 ± 2.5 | 34 ± 9 | 18 ± 7 | 22 ± 7 | 41 | 69 |
| Creatine | 580 ± 80 | 630 ± 100 | 750 ± 80 | 1000 ± 200 | 1100 ± 100 | 2800 ± 400 | 730 ± 80 | 1400 ± 100 | 780 ± 100 | 1100 ± 200 | 810 ± 90 | 830 ± 110 | 3600 | 2300 |
| Ergothioneine | 1900 ± 800 | 1900 ± 700 | 4500 ± 900 | 3000 ± 500 | 4100 ± 600 | 9100 ± 1400 | 3800 ± 600 | 3700 ± 600 | 3200 ± 200 | 2900 ± 500 | 1600 ± 300 | 3600 ± 500 | 1000 | 360 |
| Formate | 75 ± 11 | 240 ± 30 | 83 ± 33 | 320 ± 80 | 130 ± 30 | 320 ± 60 | 220 ± 110 | 180 ± 80 | 68 ± 12 | 130 ± 20 | 84 ± 52 | 230 ± 50 | 150 | 100 |
| Fumarate | 13 ± 4 | 23 ± 9 | 19 ± 4 | 36 ± 8 | 29 ± 6 | 24 ± 6 | 32 ± 10 | 10 ± 4 | 18 ± 2 | 24 ± 4 | 25 ± 7 | 14 ± 5 | 27 | 26 |
| Gl-Ph-Choline | 53 ± 25 | 300 ± 30 | 390 ± 60 | 20 ± 15 | 230 ± 50 | 240 ± 120 | 270 ± 40 | 140 ± 30 | 400 ± 20 | 140 ± 30 | 0 | 240 ± 40 | 140 | 41 |
| Glucose | 270 ± 200 | 1400 ± 300 | 820 ± 390 | 1400 ± 200 | 970 ± 230 | 500 ± 240 | 1600 ± 400 | 1900 ± 200 | 950 ± 200 | 450 ± 230 | 1800 ± 500 | 800 ± 310 | 960 | 0 |
| Glutamate | 790 ± 70 | 1800 ± 300 | 1900 ± 400 | 1600 ± 300 | 3900 ± 400 | 1000 ± 100 | 2700 ± 300 | 3000 ± 100 | 2100 ± 300 | 2800 ± 300 | 2000 ± 300 | 1900 ± 100 | 1300 | 1600 |
| Glutamine | 1700 ± 300 | 5800 ± 1400 | 5400 ± 800 | 4400 ± 1400 | 7800 ± 1600 | 4600 ± 1700 | 11,000 ± 1000 | 11,000 ± 1000 | 6400 ± 300 | 9200 ± 1100 | 3200 ± 500 | 7400 ± 1800 | 6100 | 2400 |
| Glutathione | 1200 ± 500 | 3500 ± 600 | 2900 ± 300 | 3900 ± 200 | 4000 ± 400 | 6000 ± 1000 | 4700 ± 300 | 4600 ± 100 | 3600 ± 700 | 4400 ± 1200 | 2100 ± 400 | 3600 ± 600 | 1800 | 1400 |
| Glycerol | 220 ± 80 | 120 ± 100 | 99 ± 66 | 260 ± 300 | 0 | 240 ± 80 | 0 | 0 | 71 ± 37 | 0 | 0 | 70 ± 44 | 0 | 0 |
| Glycine | 580 ± 110 | 310 ± 130 | 530 ± 140 | 440 ± 70 | 530 ± 120 | 900 ± 250 | 550 ± 100 | 950 ± 110 | 470 ± 90 | 1100 ± 100 | 350 ± 70 | 540 ± 80 | 150 | 560 |
| GSSG | 330 ± 120 | 400 ± 80 | 280 ± 50 | 380 ± 90 | 560 ± 150 | 350 ± 40 | 500 ± 190 | 350 ± 60 | 260 ± 60 | 940 ± 530 | 0 | 280 ± 40 | 0 | 0 |
| GTP | 120 ± 40 | 0 | 270 ± 30 | 0 | 150 ± 20 | 220 ± 30 | 260 ± 80 | 110 ± 40 | 0 | 210 ± 50 | 190 ± 70 | 270 ± 50 | 320 | 200 |
| Histidine | 120 ± 30 | 97 ± 12 | 90 ± 24 | 110 ± 50 | 140 ± 20 | 500 ± 100 | 210 ± 40 | 430 ± 80 | 140 ± 50 | 310 ± 50 | 85 ± 34 | 100 ± 40 | 190 | 98 |
| Hypoxanthine | 250 ± 40 | 94 ± 17 | 50 ± 27 | 81 ± 11 | 51 ± 8 | 130 ± 10 | 84 ± 20 | 53 ± 12 | 66 ± 15 | 140 ± 50 | 180 ± 30 | 73 ± 15 | 130 | 240 |
| Inosinate | 26 ± 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 42 ± 23 | 15 ± 26 | 0 | 11 | 58 |
| Inosine | 89 ± 13 | 17 ± 14 | 27 ± 28 | 24 ± 19 | 34 ± 3 | 52 ± 14 | 42 ± 11 | 29 ± 1 | 18 ± 13 | 66 ± 14 | 7.7 ± 13.9 | 44 ± 7 | 0 | 0 |
| Isobutyrate | 8 ± 4.9 | 7.3 ± 1 | 4.6 ± 2.3 | 15 ± 7 | 8.7 ± 2.3 | 9.6 ± 4.3 | 10 ± 5 | 7 ± 3.6 | 8 ± 2.6 | 16 ± 5 | 1.5 ± 2.5 | 4.2 ± 2.8 | 19 | 11 |
| Isoleucine | 82 ± 20 | 34 ± 5 | 34 ± 11 | 73 ± 15 | 24 ± 6 | 78 ± 14 | 22 ± 5 | 29 ± 10 | 42 ± 5 | 46 ± 9 | 36 ± 9 | 40 ± 9 | 240 | 320 |
| Lactate | 14,000 ± 2000 | 7100 ± 1100 | 7600 ± 3300 | 5500 ± 1100 | 6400 ± 500 | 7700 ± 1600 | 8100 ± 1100 | 6100 ± 200 | 5300 ± 700 | 12,000 ± 2000 | 7100 ± 700 | 4900 ± 800 | 21,000 | 6700 |
| Leucine | 190 ± 40 | 110 ± 10 | 140 ± 40 | 160 ± 30 | 98 ± 18 | 2200 ± 400 | 130 ± 30 | 100 ± 20 | 160 ± 30 | 170 ± 20 | 85 ± 13 | 120 ± 20 | 950 | 740 |
| Lysine | 140 ± 40 | 67 ± 23 | 62 ± 26 | 300 ± 50 | 0 | 910 ± 140 | 0 | 0 | 65 ± 30 | 0 | 12 ± 30 | 77 ± 18 | 160 | 88 |
| Methionine | 230 ± 50 | 430 ± 170 | 960 ± 240 | 1000 ± 200 | 590 ± 110 | 350 ± 130 | 470 ± 100 | 500 ± 30 | 550 ± 170 | 610 ± 80 | 200 ± 50 | 380 ± 110 | 480 | 170 |
| myo-Inositol | 38,000 ± 2000 | 29,000 ± 9000 | 26,000 ± 2000 | 29,000 ± 4000 | 29,000 ± 2000 | 28,000 ± 3000 | 29,000 ± 1000 | 25,000 ± 2000 | 26,000 ± 0 | 34,000 ± 2000 | 37,000 ± 4000 | 32,000 ± 1000 | 3000 | 5400 |
| N,N-DMG 2 | 30 ± 13 | 0 | 0 | 0 | 0 | 39 ± 8 | 0 | 0 | 0 | 0 | 0 | 72 ± 13 | 0 | 0 |
| NAA | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 270 | 500 |
| NAD | 500 ± 60 | 250 ± 50 | 150 ± 40 | 120 ± 50 | 100 ± 20 | 250 ± 30 | 130 ± 50 | 50 ± 9 | 130 ± 40 | 190 ± 70 | 220 ± 40 | 210 ± 20 | 320 | 250 |
| NADH | 1200 ± 100 | 48 ± 17 | 17 ± 10 | 86 ± 14 | 8.3 ± 3.6 | 560 ± 270 | 13 ± 7 | 7.7 ± 3.1 | 28 ± 13 | 16 ± 8 | 2.7 ± 5.3 | 0 | 0 | 9 |
| NADPH 2 | 0 | 0 | 0 | 0 | 0 | 300 ± 70 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| N-Me-His 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 200 | 80 |
| Ph-Choline | 170 ± 50 | 89 ± 59 | 550 ± 50 | 340 ± 30 | 69 ± 14 | 110 ± 20 | 64 ± 21 | 170 ± 20 | 96 ± 68 | 210 ± 20 | 250 ± 30 | 200 ± 80 | 3900 | 3900 |
| Phenylalanine | 180 ± 70 | 100 ± 50 | 50 ± 14 | 74 ± 24 | 39 ± 4 | 53 ± 15 | 24 ± 19 | 49 ± 10 | 67 ± 7 | 44 ± 16 | 41 ± 16 | 48 ± 18 | 510 | 580 |
| Proline | 480 ± 110 | 320 ± 120 | 600 ± 120 | 260 ± 60 | 1000 ± 200 | 680 ± 240 | 480 ± 120 | 580 ± 110 | 690 ± 150 | 670 ± 90 | 650 ± 350 | 560 ± 140 | 620 | 280 |
| Pyroglutamate | 560 ± 60 | 800 ± 190 | 920 ± 240 | 920 ± 180 | 1700 ± 200 | 1700 ± 200 | 1200 ± 200 | 1900 ± 200 | 1100 ± 100 | 1300 ± 200 | 650 ± 120 | 840 ± 90 | 340 | 170 |
| Pyruvate | 6.8 ± 1.3 | 9.3 ± 1 | 16 ± 4 | 12 ± 4 | 9.2 ± 3.2 | 11 ± 5 | 16 ± 6 | 7 ± 0 | 11 ± 2 | 15 ± 3 | 13 ± 3 | 6.8 ± 2.9 | 9 | 14 |
| Sarcosine 2 | 7 ± 4.4 | 4 ± 2.8 | 7.8 ± 1.9 | 15 ± 5 | 7.8 ± 2.3 | 22 ± 19 | 5.7 ± 4.1 | 6.7 ± 0.6 | 7.7 ± 2.1 | 18 ± 4 | 7.4 ± 3.9 | 9.8 ± 3.8 | 35 | 30 |
| scyllo-Inositol | 50 ± 12 | 34 ± 8 | 45 ± 11 | 51 ± 22 | 70 ± 17 | 750 ± 250 | 51 ± 10 | 39 ± 7 | 44 ± 9 | 49 ± 8 | 55 ± 18 | 54 ± 13 | 210 | 170 |
| Serine | 2700 ± 200 | 2200 ± 100 | 4400 ± 1500 | 2200 ± 700 | 3900 ± 400 | 1200 ± 300 | 4600 ± 500 | 3100 ± 300 | 3200 ± 600 | 2900 ± 800 | 900 ± 240 | 2800 ± 500 | 5600 | 2900 |
| Taurine | 15,000 ± 3000 | 20,000 ± 4000 | 13,000 ± 1000 | 20,000 ± 4000 | 35,000 ± 2000 | 3200 ± 500 | 38,000 ± 5000 | 48,000 ± 3000 | 15,000 ± 3000 | 36,000 ± 2000 | 15,000 ± 2000 | 14,000 ± 1000 | 32,000 | 26,000 |
| Threonine | 300 ± 80 | 300 ± 150 | 420 ± 120 | 820 ± 180 | 440 ± 90 | 860 ± 30 | 500 ± 80 | 370 ± 80 | 480 ± 110 | 540 ± 40 | 260 ± 100 | 220 ± 70 | 540 | 120 |
| Tryptophan | 71 ± 38 | 95 ± 44 | 31 ± 17 | 65 ± 12 | 0 | 0 | 0 | 0 | 37 ± 6 | 0 | 0 | 0 | 160 | 130 |
| Tyrosine | 200 ± 50 | 200 ± 80 | 200 ± 40 | 180 ± 60 | 130 ± 40 | 130 ± 30 | 230 ± 70 | 170 ± 10 | 210 ± 20 | 170 ± 40 | 140 ± 40 | 170 ± 60 | 260 | 97 |
| UDP | 330 ± 80 | 310 ± 60 | 300 ± 50 | 410 ± 60 | 260 ± 60 | 490 ± 30 | 340 ± 60 | 210 ± 20 | 270 ± 60 | 310 ± 50 | 150 ± 60 | 280 ± 50 | 1600 | 590 |
| Valine | 210 ± 30 | 68 ± 13 | 92 ± 24 | 200 ± 40 | 100 ± 20 | 190 ± 10 | 110 ± 30 | 95 ± 3 | 110 ± 30 | 170 ± 20 | 110 ± 20 | 94 ± 18 | 1100 | 1200 |
| S109 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 95 ± 23 | 0 | 0 | 0 |
| S112 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 81 ± 23 | 0 | 0 | 0 | 0 | 0 | 0 |
| S120 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 130 ± 30 | 0 | 0 | 0 | 0 | 0 | 0 |
| D121 2 | 0 | 0 | 0 | 0 | 0 | 950 ± 220 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| D139 2 | 0 | 0 | 0 | 0 | 0 | 1100 ± 500 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| T727 2 | 0 | 0 | 0 | 0 | 0 | 400 ± 180 | 0 | 0 | 0 | 0 | 0 | 36 ± 6 | 0 | 0 |
| S823 2 | 100 ± 40 | 0 | 0 | 0 | 0 | 670 ± 160 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zelentsova, E.A.; Yanshole, L.V.; Tsentalovich, Y.P.; Sharshov, K.A.; Yanshole, V.V. The Application of Quantitative Metabolomics for the Taxonomic Differentiation of Birds. Biology 2022, 11, 1089. https://doi.org/10.3390/biology11071089
Zelentsova EA, Yanshole LV, Tsentalovich YP, Sharshov KA, Yanshole VV. The Application of Quantitative Metabolomics for the Taxonomic Differentiation of Birds. Biology. 2022; 11(7):1089. https://doi.org/10.3390/biology11071089
Chicago/Turabian StyleZelentsova, Ekaterina A., Lyudmila V. Yanshole, Yuri P. Tsentalovich, Kirill A. Sharshov, and Vadim V. Yanshole. 2022. "The Application of Quantitative Metabolomics for the Taxonomic Differentiation of Birds" Biology 11, no. 7: 1089. https://doi.org/10.3390/biology11071089
APA StyleZelentsova, E. A., Yanshole, L. V., Tsentalovich, Y. P., Sharshov, K. A., & Yanshole, V. V. (2022). The Application of Quantitative Metabolomics for the Taxonomic Differentiation of Birds. Biology, 11(7), 1089. https://doi.org/10.3390/biology11071089