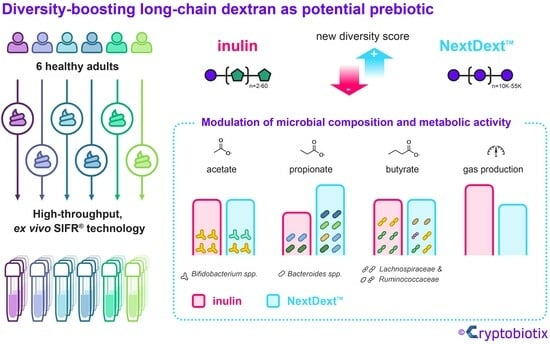
A Long-Chain Dextran Produced by Weissella cibaria Boosts the Diversity of Health-Related Gut Microbes Ex Vivo
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Test Compounds
2.2. SIFR® Technology
2.3. Key Fermentation Parameters
2.4. Microbiota Phylogenetic Analysis: Quantitative 16S rRNA Gene Profiling
2.5. Diversity Indices
2.6. Statistical Analysis
3. Results
3.1. Microbiota of Six Human Adults Cover Clinically Relevant Interpersonal Differences
3.2. Dextran Stimulated the Growth of Human Adult Gut Microbiota Ex Vivo
3.3. Dextran Exhibited Prebiotic Effects on Species Richness and Evenness of the Gut Microbiota According to Traditional α-Diversity Indices
3.4. Considerations on Limitations and Interpretation of Outcomes of Traditional Diversity Indices
3.5. The Novel Community Modulation Score Shows That Dextran Supported a High Microbial Diversity
3.6. Dextran Was Selectively Fermented by a Broad Spectrum of Human Gut Microbes Ex Vivo
3.7. Dextran Similarly Boosted Production of Health-Related SCFA While Inducing Less Gas Production Than IN
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Afzaal, M.; Saeed, F.; Shah, Y.A.; Hussain, M.; Rabail, R.; Socol, C.T.; Hassoun, A.; Pateiro, M.; Lorenzo, J.M.; Rusu, A.V.; et al. Human Gut Microbiota in Health and Disease: Unveiling the Relationship. Front. Microbiol. 2022, 13, 999001. [Google Scholar] [CrossRef]
- Paone, P.; Cani, P.D. Mucus Barrier, Mucins and Gut Microbiota: The Expected Slimy Partners? Gut 2020, 69, 2232–2243. [Google Scholar] [CrossRef]
- Peron, G.; Gargari, G.; Meroño, T.; Miñarro, A.; Lozano, E.V.; Escuder, P.C.; González-Domínguez, R.; Hidalgo-Liberona, N.; Del Bo’, C.; Bernardi, S.; et al. Crosstalk among Intestinal Barrier, Gut Microbiota and Serum Metabolome after a Polyphenol-Rich Diet in Older Subjects with “Leaky Gut”: The MaPLE Trial. Clin. Nutr. 2021, 40, 5288–5297. [Google Scholar] [CrossRef]
- Connors, J.; Dunn, K.A.; Allott, J.; Bandsma, R.; Rashid, M.; Otley, A.R.; Bielawski, J.P.; Van Limbergen, J. The Relationship between Fecal Bile Acids and Microbiome Community Structure in Pediatric Crohn’s Disease. ISME J. 2020, 14, 702–713. [Google Scholar] [CrossRef]
- de la Fuente-Nunez, C.; Meneguetti, B.T.; Franco, O.L.; Lu, T.K. Neuromicrobiology: How Microbes Influence the Brain. ACS Chem. Neurosci. 2018, 9, 141–150. [Google Scholar] [CrossRef]
- Zheng, D.; Liwinski, T.; Elinav, E. Interaction between Microbiota and Immunity in Health and Disease. Cell Res. 2020, 30, 492–506. [Google Scholar] [CrossRef]
- Krautkramer, K.A.; Fan, J.; Bäckhed, F. Gut Microbial Metabolites as Multi-Kingdom Intermediates. Nat. Rev. Microbiol. 2021, 19, 77–94. [Google Scholar] [CrossRef]
- Koh, A.; De Vadder, F.; Kovatcheva-Datchary, P.; Bäckhed, F. From Dietary Fiber to Host Physiology: Short-Chain Fatty Acids as Key Bacterial Metabolites. Cell 2016, 165, 1332–1345. [Google Scholar] [CrossRef]
- Louis, P.; Flint, H.J. Formation of Propionate and Butyrate by the Human Colonic Microbiota. Environ. Microbiol. 2017, 19, 29–41. [Google Scholar] [CrossRef]
- Morrison, D.J.; Preston, T. Formation of Short Chain Fatty Acids by the Gut Microbiota and Their Impact on Human Metabolism. Gut Microbes 2016, 7, 189–200. [Google Scholar] [CrossRef]
- Belenguer, A.; Duncan, S.H.; Calder, A.G.; Holtrop, G.; Louis, P.; Lobley, G.E.; Flint, H.J. Two Routes of Metabolic Cross-Feeding between Bifidobacterium adolescentis and Butyrate-Producing Anaerobes from the Human Gut. Appl. Environ. Microbiol. 2006, 72, 3593–3599. [Google Scholar] [CrossRef]
- Bunesova, V.; Lacroix, C.; Schwab, C. Mucin Cross-Feeding of Infant Bifidobacteria and Eubacterium hallii. Microb. Ecol. 2018, 75, 228–238. [Google Scholar] [CrossRef]
- Rios-Covian, D.; Gueimonde, M.; Duncan, S.H.; Flint, H.J.; de los Reyes-Gavilan, C.G. Enhanced Butyrate Formation by Cross-Feeding between Faecalibacterium prausnitzii and Bifidobacterium adolescentis. FEMS Microbiol. Lett. 2015, 362, fnv176. [Google Scholar] [CrossRef]
- Davani-Davari, D.; Negahdaripour, M.; Karimzadeh, I.; Seifan, M.; Mohkam, M.; Masoumi, S.; Berenjian, A.; Ghasemi, Y. Prebiotics: Definition, Types, Sources, Mechanisms, and Clinical Applications. Foods 2019, 8, 92. [Google Scholar] [CrossRef]
- Gibson, G.R.; Hutkins, R.; Sanders, M.E.; Prescott, S.L.; Reimer, R.A.; Salminen, S.J.; Scott, K.; Stanton, C.; Swanson, K.S.; Cani, P.D.; et al. Expert Consensus Document: The International Scientific Association for Probiotics and Prebiotics (ISAPP) Consensus Statement on the Definition and Scope of Prebiotics. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 491–502. [Google Scholar] [CrossRef]
- Gotteland, M.; Riveros, K.; Gasaly, N.; Carcamo, C.; Magne, F.; Liabeuf, G.; Beattie, A.; Rosenfeld, S. The Pros and Cons of Using Algal Polysaccharides as Prebiotics. Front. Nutr. 2020, 7, 163. [Google Scholar] [CrossRef]
- Kaur, A.P.; Bhardwaj, S.; Dhanjal, D.S.; Nepovimova, E.; Cruz-Martins, N.; Kuča, K.; Chopra, C.; Singh, R.; Kumar, H.; Șen, F.; et al. Plant Prebiotics and Their Role in the Amelioration of Diseases. Biomolecules 2021, 11, 440. [Google Scholar] [CrossRef]
- Guan, Z.; Feng, Q. Chitosan and Chitooligosaccharide: The Promising Non-Plant-Derived Prebiotics with Multiple Biological Activities. Int. J. Mol. Sci. 2022, 23, 6761. [Google Scholar] [CrossRef]
- Khan, R.; Shah, M.D.; Shah, L.; Lee, P.-C.; Khan, I. Bacterial Polysaccharides—A Big Source for Prebiotics and Therapeutics. Front. Nutr. 2022, 9, 1031935. [Google Scholar] [CrossRef]
- Amaretti, A.; Bottari, B.; Morreale, F.; Savo Sardaro, M.L.; Angelino, D.; Raimondi, S.; Rossi, M.; Pellegrini, N. Potential Prebiotic Effect of a Long-Chain Dextran Produced by Weissella cibaria: An In Vitro Evaluation. Int. J. Food Sci. Nutr. 2020, 71, 563–571. [Google Scholar] [CrossRef]
- Baruah, R.; Maina, N.H.; Katina, K.; Juvonen, R.; Goyal, A. Functional Food Applications of Dextran from Weissella cibaria RBA12 from Pummelo (Citrus maxima). Int. J. Food Microbiol. 2017, 242, 124–131. [Google Scholar] [CrossRef]
- Sarbini, S.R.; Kolida, S.; Deaville, E.R.; Gibson, G.R.; Rastall, R.A. Potential of Novel Dextran Oligosaccharides as Prebiotics for Obesity Management through In Vitro Experimentation. Br. J. Nutr. 2014, 112, 1303–1314. [Google Scholar] [CrossRef]
- Kim, G.; Bae, J.-H.; Cheon, S.; Lee, D.H.; Kim, D.H.; Lee, D.; Park, S.-H.; Shim, S.; Seo, J.-H.; Han, N.S. Prebiotic Activities of Dextran from Leuconostoc mesenteroides SPCL742 Analyzed in the Aspect of the Human Gut Microbial Ecosystem. Food Funct. 2022, 13, 1256–1267. [Google Scholar] [CrossRef]
- Díaz-Montes, E. Dextran: Sources, Structures, and Properties. Polysaccharides 2021, 2, 554–565. [Google Scholar] [CrossRef]
- Le Bastard, Q.; Chapelet, G.; Javaudin, F.; Lepelletier, D.; Batard, E.; Montassier, E. The Effects of Inulin on Gut Microbial Composition: A Systematic Review of Evidence from Human Studies. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 403–413. [Google Scholar] [CrossRef]
- Costa, J.; Ahluwalia, A. Advances and Current Challenges in Intestinal in Vitro Model Engineering: A Digest. Front. Bioeng. Biotechnol. 2019, 7, 144. [Google Scholar] [CrossRef]
- Procházková, N.; Falony, G.; Dragsted, L.O.; Licht, T.R.; Raes, J.; Roager, H.M. Advancing Human Gut Microbiota Research by Considering Gut Transit Time. Gut 2022, 72, 180–191. [Google Scholar] [CrossRef]
- O’Donnell, M.M.; Rea, M.C.; Shanahan, F.; Ross, R.P. The Use of a Mini-Bioreactor Fermentation System as a Reproducible, High-Throughput Ex Vivo Batch Model of the Distal Colon. Front. Microbiol. 2018, 9, 1844. [Google Scholar] [CrossRef]
- Van den Abbeele, P.; Verstrepen, L.; Ghyselinck, J.; Albers, R.; Marzorati, M.; Mercenier, A. A Novel Non-Digestible, Carrot-Derived Polysaccharide (cRG-I) Selectively Modulates the Human Gut Microbiota While Promoting Gut Barrier Integrity: An Integrated In Vitro Approach. Nutrients 2020, 12, 1917. [Google Scholar] [CrossRef]
- Biagini, F.; Calvigioni, M.; Lapomarda, A.; Vecchione, A.; Magliaro, C.; De Maria, C.; Montemurro, F.; Celandroni, F.; Mazzantini, D.; Mattioli-Belmonte, M.; et al. A Novel 3D in Vitro Model of the Human Gut Microbiota. Sci. Rep. 2020, 10, 21499. [Google Scholar] [CrossRef]
- Gaisawat, M.B.; MacPherson, C.W.; Tremblay, J.; Piano, A.; Iskandar, M.M.; Tompkins, T.A.; Kubow, S. Probiotic Supplementation in a Clostridium Difficile-Infected Gastrointestinal Model Is Associated with Restoring Metabolic Function of Microbiota. Microorganisms 2019, 8, 60. [Google Scholar] [CrossRef]
- Rajilić-Stojanović, M.; Maathuis, A.; Heilig, H.G.H.J.; Venema, K.; de Vos, W.M.; Smidt, H. Evaluating the Microbial Diversity of an In Vitro Model of the Human Large Intestine by Phylogenetic Microarray Analysis. Microbiology 2010, 156, 3270–3281. [Google Scholar] [CrossRef]
- Van den Abbeele, P.; Grootaert, C.; Marzorati, M.; Possemiers, S.; Verstraete, W.; Gérard, P.; Rabot, S.; Bruneau, A.; El Aidy, S.; Derrien, M.; et al. Microbial Community Development in a Dynamic Gut Model Is Reproducible, Colon Region Specific, and Selective for Bacteroidetes and Clostridium Cluster IX. Appl. Environ. Microbiol. 2010, 76, 5237–5246. [Google Scholar] [CrossRef]
- Van den Abbeele, P.; Deyaert, S.; Thabuis, C.; Perreau, C.; Bajic, D.; Wintergerst, E.; Joossens, M.; Firrman, J.; Walsh, D.; Baudot, A. Bridging Preclinical and Clinical Gut Microbiota Research Using the Ex Vivo SIFR® Technology. Front. Microbiol. 2023, 14, 1131662. [Google Scholar] [CrossRef]
- Falony, G.; Joossens, M.; Vieira-Silva, S.; Wang, J.; Darzi, Y.; Faust, K.; Kurilshikov, A.; Bonder, M.J.; Valles-Colomer, M.; Vandeputte, D.; et al. Population-Level Analysis of Gut Microbiome Variation. Science 2016, 352, 560–564. [Google Scholar] [CrossRef]
- Husson, F.; Josse, J.; Le, S.; Mazet, J. FactoMineR: Multivariate Exploratory Data Analysis and Data Mining; 2022. Available online: https://cran.r-project.org/web/packages/FactoMineR/index.html (accessed on 28 October 2023).
- Rohart, F.; Gautier, B.; Singh, A.; Cao, K.-A.L. mixOmics: An R Package for ‘omics Feature Selection and Multiple Data Integration. PLoS Comput. Biol. 2017, 13, e1005752. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Kassambara, A. Rstatix: Pipe-Friendly Framework for Basic Statistical Tests. 2022. Available online: https://cran.r-project.org/web/packages/rstatix/index.html#:~:text=rstatix%3A%20Pipe%2DFriendly%20Framework%20for,Kruskal%2DWallis%20and%20correlation%20analyses. (accessed on 28 October 2023).
- Wickham, H.; Chang, W.; Henry, L.; Pedersen, T.L.; Takahashi, K.; Wilke, C.; Woo, K.; Yutani, H.; Dunnington, D. Ggplot2: Create Elegant Data Visualisations Using the Grammar of Graphics. 2022. Available online: https://cran.r-project.org/web/packages/ggplot2/index.html (accessed on 28 October 2023).
- Costea, P.I.; Hildebrand, F.; Arumugam, M.; Bäckhed, F.; Blaser, M.J.; Bushman, F.D.; de Vos, W.M.; Ehrlich, S.D.; Fraser, C.M.; Hattori, M.; et al. Enterotypes in the Landscape of Gut Microbial Community Composition. Nat. Microbiol. 2018, 3, 8–16. [Google Scholar] [CrossRef]
- Devika, N.T.; Raman, K. Deciphering the Metabolic Capabilities of Bifidobacteria Using Genome-Scale Metabolic Models. Sci. Rep. 2019, 9, 18222. [Google Scholar] [CrossRef]
- Fukuda, S.; Toh, H.; Hase, K.; Oshima, K.; Nakanishi, Y.; Yoshimura, K.; Tobe, T.; Clarke, J.M.; Topping, D.L.; Suzuki, T.; et al. Bifidobacteria Can Protect from Enteropathogenic Infection through Production of Acetate. Nature 2011, 469, 543–547. [Google Scholar] [CrossRef]
- Shetty, S.A.; Zuffa, S.; Bui, T.P.N.; Aalvink, S.; Smidt, H.; De Vos, W.M. Reclassification of Eubacterium hallii as Anaerobutyricum hallii Gen. Nov., Comb. Nov., and Description of Anaerobutyricum soehngenii Sp. Nov., a Butyrate and Propionate-Producing Bacterium from Infant Faeces. Int. J. Syst. Evol. Microbiol. 2018, 68, 3741–3746. [Google Scholar] [CrossRef]
- Duncan, S.H.; Hold, G.L.; Harmsen, H.J.M.; Stewart, C.S.; Flint, H.J. Growth Requirements and Fermentation Products of Fusobacterium prausnitzii, and a Proposal to Reclassify It as Faecalibacterium prausnitzii gen. nov., comb. nov. Int. J. Syst. Evol. Microbiol. 2002, 52, 2141–2146. [Google Scholar] [CrossRef]
- Arumugam, M.; Raes, J.; Pelletier, E.; Le Paslier, D.; Yamada, T.; Mende, D.R.; Fernandes, G.R.; Tap, J.; Bruls, T.; Batto, J.-M.; et al. Enterotypes of the Human Gut Microbiome. Nature 2011, 473, 174–180. [Google Scholar] [CrossRef]
- Bindels, L.B.; Delzenne, N.M.; Cani, P.D.; Walter, J. Towards a More Comprehensive Concept for Prebiotics. Nat. Rev. Gastroenterol. Hepatol. 2015, 12, 303–310. [Google Scholar] [CrossRef]
- Gibson, G.R. Fibre and Effects on Probiotics (the Prebiotic Concept). Clin. Nutr. Suppl. 2004, 1, 25–31. [Google Scholar] [CrossRef]
- Marzorati, M.; Bubeck, S.; Bayne, T.; Krishnan, K.; Young, A. Evaluation of the Effect of Food Products Containing Prebiotics and Bacillus Subtilis HU58 on the Gut Microbial Community Activity and Community Composition Using an In Vitro M-SHIME® Model. Appl. Sci. 2021, 11, 11963. [Google Scholar] [CrossRef]
- Zhang, C.; Yin, A.; Li, H.; Wang, R.; Wu, G.; Shen, J.; Zhang, M.; Wang, L.; Hou, Y.; Ouyang, H.; et al. Dietary Modulation of Gut Microbiota Contributes to Alleviation of Both Genetic and Simple Obesity in Children. EBioMedicine 2015, 2, 968–984. [Google Scholar] [CrossRef]
- McNulty, N.P.; Wu, M.; Erickson, A.R.; Pan, C.; Erickson, B.K.; Martens, E.C.; Pudlo, N.A.; Muegge, B.D.; Henrissat, B.; Hettich, R.L.; et al. Effects of Diet on Resource Utilization by a Model Human Gut Microbiota Containing Bacteroides Cellulosilyticus WH2, a Symbiont with an Extensive Glycobiome. PLoS Biol. 2013, 11, e1001637. [Google Scholar] [CrossRef]
- Wu, M.; McNulty, N.P.; Rodionov, D.A.; Khoroshkin, M.S.; Griffin, N.W.; Cheng, J.; Latreille, P.; Kerstetter, R.A.; Terrapon, N.; Henrissat, B.; et al. Genetic Determinants of In Vivo Fitness and Diet Responsiveness in Multiple Human Gut Bacteroides. Science 2015, 350, aac5992. [Google Scholar] [CrossRef]
- Rios-Covian, D.; Salazar, N.; Gueimonde, M.; De Los Reyes-Gavilan, C.G. Shaping the Metabolism of Intestinal Bacteroides Population through Diet to Improve Human Health. Front. Microbiol. 2017, 8, 376. [Google Scholar] [CrossRef]
- Yoshida, N.; Emoto, T.; Yamashita, T.; Watanabe, H.; Hayashi, T.; Tabata, T.; Hoshi, N.; Hatano, N.; Ozawa, G.; Sasaki, N.; et al. Bacteroides Vulgatus and Bacteroides Dorei Reduce Gut Microbial Lipopolysaccharide Production and Inhibit Atherosclerosis. Circulation 2018, 138, 2486–2498. [Google Scholar] [CrossRef]
- Xu, M.; Lan, R.; Qiao, L.; Lin, X.; Hu, D.; Zhang, S.; Yang, J.; Zhou, J.; Ren, Z.; Li, X.; et al. Bacteroides Vulgatus Ameliorates Lipid Metabolic Disorders and Modulates Gut Microbial Composition in Hyperlipidemic Rats. Microbiol. Spectr. 2023, 11, e02517-22. [Google Scholar] [CrossRef]
- Todesco, T.; Rao, A.V.; Bosello, O.; Jenkins, D.J. Propionate Lowers Blood Glucose and Alters Lipid Metabolism in Healthy Subjects. Am. J. Clin. Nutr. 1991, 54, 860–865. [Google Scholar] [CrossRef]
- Hosseini, E.; Grootaert, C.; Verstraete, W.; Van de Wiele, T. Propionate as a Health-Promoting Microbial Metabolite in the Human Gut. Nutr. Rev. 2011, 69, 245–258. [Google Scholar] [CrossRef]
- Osto, E. The Promise of the Gut Metabolite Propionate for a Novel and Personalized Lipid-Lowering Treatment. Eur. Heart J. 2022, 43, 534–537. [Google Scholar] [CrossRef]
- Wang, K.; Liao, M.; Zhou, N.; Bao, L.; Ma, K.; Zheng, Z.; Wang, Y.; Liu, C.; Wang, W.; Wang, J.; et al. Parabacteroides distasonis Alleviates Obesity and Metabolic Dysfunctions via Production of Succinate and Secondary Bile Acids. Cell Rep. 2019, 26, 222–235.e5. [Google Scholar] [CrossRef]
- Chen, J.; Chen, X.; Ho, C.L. Recent Development of Probiotic bifidobacteria for Treating Human Diseases. Front. Bioeng. Biotechnol. 2021, 9, 770248. [Google Scholar]
- Lin, C.; Lin, Y.; Zhang, H.; Wang, G.; Zhao, J.; Zhang, H.; Chen, W. Intestinal ‘Infant-Type’ Bifidobacteria Mediate Immune System Development in the First 1000 Days of Life. Nutrients 2022, 14, 1498. [Google Scholar] [CrossRef]
- Derrien, M.; Turroni, F.; Ventura, M.; van Sinderen, D. Insights into Endogenous bifidobacterium Species in the Human Gut Microbiota during Adulthood. Trends Microbiol. 2022, 30, 940–947. [Google Scholar] [CrossRef]
- Alessandri, G.; Ossiprandi, M.C.; MacSharry, J.; van Sinderen, D.; Ventura, M. Bifidobacterial Dialogue with Its Human Host and Consequent Modulation of the Immune System. Front. Immunol. 2019, 10, 2348. [Google Scholar] [CrossRef] [PubMed]
- Rivière, A.; Selak, M.; Lantin, D.; Leroy, F.; De Vuyst, L. Bifidobacteria and Butyrate-Producing Colon Bacteria: Importance and Strategies for Their Stimulation in the Human Gut. Front. Microbiol. 2016, 7, 979. [Google Scholar] [CrossRef]
- Maturana, J.L.; Cárdenas, J.P. Insights on the Evolutionary Genomics of the Blautia Genus: Potential New Species and Genetic Content Among Lineages. Front. Microbiol. 2021, 12, 660920. [Google Scholar] [CrossRef] [PubMed]
- Gossling, J.; Moore, W.E.C. Gemmiger formicilis, n.gen., n.sp., an Anaerobic Budding Bacterium from Intestines. Int. J. Syst. Evol. Microbiol. 1975, 25, 202–207. [Google Scholar] [CrossRef]
- Peng, L.; Li, Z.-R.; Green, R.S.; Holzmanr, I.R.; Lin, J. Butyrate Enhances the Intestinal Barrier by Facilitating Tight Junction Assembly via Activation of AMP-Activated Protein Kinase in Caco-2 Cell Monolayers. J. Nutr. 2009, 139, 1619–1625. [Google Scholar] [CrossRef] [PubMed]
- Plöger, S.; Stumpff, F.; Penner, G.B.; Schulzke, J.-D.; Gäbel, G.; Martens, H.; Shen, Z.; Günzel, D.; Aschenbach, J.R. Microbial Butyrate and Its Role for Barrier Function in the Gastrointestinal Tract: Butyrate and the Gastrointestinal Barrier. Ann. N. Y. Acad. Sci. 2012, 1258, 52–59. [Google Scholar] [CrossRef]
- VanHook, A.M. Butyrate Benefits the Intestinal Barrier. Sci. Signal. 2015, 8, ec135. [Google Scholar] [CrossRef]
- Gao, Z.; Yin, J.; Zhang, J.; Ward, R.E.; Martin, R.J.; Lefevre, M.; Cefalu, W.T.; Ye, J. Butyrate Improves Insulin Sensitivity and Increases Energy Expenditure in Mice. Diabetes 2009, 58, 1509–1517. [Google Scholar] [CrossRef]
- Udayappan, S.; Manneras-Holm, L.; Chaplin-Scott, A.; Belzer, C.; Herrema, H.; Dallinga-Thie, G.M.; Duncan, S.H.; Stroes, E.S.G.; Groen, A.K.; Flint, H.J.; et al. Oral Treatment with Eubacterium hallii Improves Insulin Sensitivity in Db/Db Mice. npj Biofilms Microbiomes 2016, 2, 16009. [Google Scholar] [CrossRef]
- Van Deuren, T.; Blaak, E.E.; Canfora, E.E. Butyrate to Combat Obesity and Obesity-associated Metabolic Disorders: Current Status and Future Implications for Therapeutic Use. Obes. Rev. 2022, 23, e13498. [Google Scholar] [CrossRef]
- Parsaei, M.; Sarafraz, N.; Moaddab, S.Y.; Ebrahimzadeh Leylabadlo, H. The Importance of Faecalibacterium prausnitzii in Human Health and Diseases. New Microbes New Infect. 2021, 43, 100928. [Google Scholar] [CrossRef]
- Lopez-Siles, M.; Duncan, S.H.; Garcia-Gil, L.J.; Martinez-Medina, M. Faecalibacterium prausnitzii: From Microbiology to Diagnostics and Prognostics. ISME J. 2017, 11, 841–852. [Google Scholar] [CrossRef] [PubMed]
- Cuffaro, B.; Assohoun, A.L.W.; Boutillier, D.; Súkeníková, L.; Desramaut, J.; Boudebbouze, S.; Salomé-Desnoulez, S.; Hrdý, J.; Waligora-Dupriet, A.-J.; Maguin, E.; et al. In Vitro Characterization of Gut Microbiota-Derived Commensal Strains: Selection of Parabacteroides Distasonis Strains Alleviating TNBS-Induced Colitis in Mice. Cells 2020, 9, 2104. [Google Scholar] [CrossRef]
- Ezeji, J.C.; Sarikonda, D.K.; Hopperton, A.; Erkkila, H.L.; Cohen, D.E.; Martinez, S.P.; Cominelli, F.; Kuwahara, T.; Dichosa, A.E.K.; Good, C.E.; et al. Parabacteroides Distasonis: Intriguing Aerotolerant Gut Anaerobe with Emerging Antimicrobial Resistance and Pathogenic and Probiotic Roles in Human Health. Gut Microbes 2021, 13, 1922241. [Google Scholar] [CrossRef]
- Sun, H.; Guo, Y.; Wang, H.; Yin, A.; Hu, J.; Yuan, T.; Zhou, S.; Xu, W.; Wei, P.; Yin, S.; et al. Gut Commensal Parabacteroides distasonis Alleviates Inflammatory Arthritis. Gut 2023, 72, 1664–1677. [Google Scholar] [CrossRef]
- Yu, X.; Gurry, T.; Nguyen, L.T.T.; Richardson, H.S.; Alm, E.J. Prebiotics and Community Composition Influence Gas Production of the Human Gut Microbiota. mBio 2020, 11, e00217-20. [Google Scholar] [CrossRef]
- Livesey, G. Tolerance of Low-Digestible Carbohydrates: A General View. Br. J. Nutr. 2001, 85, S7–S16. [Google Scholar] [CrossRef] [PubMed]
- Marteau, P.; Seksik, P. Tolerance of Probiotics and Prebiotics. J. Clin. Gastroenterol. 2004, 38, S67–S69. [Google Scholar] [CrossRef]
- Smiricky-Tjardes, M.R.; Flickinger, E.A.; Grieshop, C.M.; Bauer, L.L.; Murphy, M.R.; Fahey, G.C. In Vitro Fermentation Characteristics of Selected Oligosaccharides by Swine Fecal Microflora. J. Anim. Sci. 2003, 81, 2505–2514. [Google Scholar] [CrossRef] [PubMed]
- Mutuyemungu, E.; Singh, M.; Liu, S.; Rose, D.J. Intestinal Gas Production by the Gut Microbiota: A Review. J. Funct. Foods 2023, 100, 105367. [Google Scholar] [CrossRef]
- Hinnebusch, B.F.; Meng, S.; Wu, J.T.; Archer, S.Y.; Hodin, R.A. The Effects of Short-Chain Fatty Acids on Human Colon Cancer Cell Phenotype Are Associated with Histone Hyperacetylation. J. Nutr. 2002, 132, 1012–1017. [Google Scholar] [CrossRef]
- McDonald, J.A.K.; Mullish, B.H.; Pechlivanis, A.; Liu, Z.; Brignardello, J.; Kao, D.; Holmes, E.; Li, J.V.; Clarke, T.B.; Thursz, M.R.; et al. Inhibiting Growth of Clostridioides Difficile by Restoring Valerate, Produced by the Intestinal Microbiota. Gastroenterology 2018, 155, 1495–1507.e15. [Google Scholar] [CrossRef] [PubMed]







Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tintoré, M.; Cuñé, J.; Vu, L.D.; Poppe, J.; Van den Abbeele, P.; Baudot, A.; de Lecea, C. A Long-Chain Dextran Produced by Weissella cibaria Boosts the Diversity of Health-Related Gut Microbes Ex Vivo. Biology 2024, 13, 51. https://doi.org/10.3390/biology13010051
Tintoré M, Cuñé J, Vu LD, Poppe J, Van den Abbeele P, Baudot A, de Lecea C. A Long-Chain Dextran Produced by Weissella cibaria Boosts the Diversity of Health-Related Gut Microbes Ex Vivo. Biology. 2024; 13(1):51. https://doi.org/10.3390/biology13010051
Chicago/Turabian StyleTintoré, Maria, Jordi Cuñé, Lam Dai Vu, Jonas Poppe, Pieter Van den Abbeele, Aurélien Baudot, and Carlos de Lecea. 2024. "A Long-Chain Dextran Produced by Weissella cibaria Boosts the Diversity of Health-Related Gut Microbes Ex Vivo" Biology 13, no. 1: 51. https://doi.org/10.3390/biology13010051
APA StyleTintoré, M., Cuñé, J., Vu, L. D., Poppe, J., Van den Abbeele, P., Baudot, A., & de Lecea, C. (2024). A Long-Chain Dextran Produced by Weissella cibaria Boosts the Diversity of Health-Related Gut Microbes Ex Vivo. Biology, 13(1), 51. https://doi.org/10.3390/biology13010051