Novel Cold-Adapted Esterase MHlip from an Antarctic Soil Metagenome
Abstract
:1. Introduction
2. Results and Discussion
2.1. MHlip Isolation and Sequence Characterization


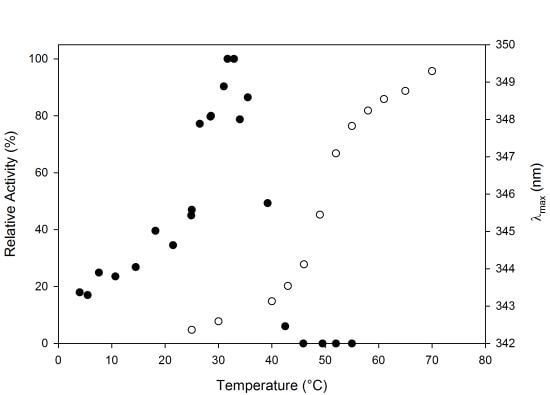
2.2. Biochemical Characterization

2.3. Thermal Stability

3. Experimental Section
3.1. Metagenomic Library Construction and Screening
3.2. Expression and Purification
3.3. Activity Assays
3.4. Thermal Stability and Unfolding
4. Conclusions

Acknowledgments
References and Notes
- Dalrymple, B.P.; Swadling, Y.; Cybinski, D.H.; Xue, G.P. Cloning of a gene encoding cinnamoyl ester hydrolase from the ruminal bacterium butyrivibrio fibrisolvens e14 by a novel method. FEMS Microbiol. Lett. 1996, 143, 115–120. [Google Scholar] [CrossRef]
- McQueen, D.A.; Schottel, J.L. Purification and characterization of a novel extracellular esterase from pathogenic streptomyces scabies that is inducible by zinc. J. Bacteriol. 1987, 169, 1967–1971. [Google Scholar]
- von der Haar, B.; Walter, S.; Schwapenheuer, S.; Schrempf, H. A novel fusidic acid resistance gene from streptomyces lividans 66 encodes a highly specific esterase. Microbiology 1997, 143, 867–874. [Google Scholar] [CrossRef]
- Schloss, P.D.; Handelsman, J. Metagenomics for studying unculturable microorganisms: Cutting the gordian knot. Genome Biol. 2005, 6, 229. [Google Scholar] [CrossRef]
- Casanueva, A.; Tuffin, M.; Cary, C.; Cowan, D.A. Molecular adaptations to psychrophily: The impact of 'omic' technologies. Trends Microbiol. 2010, 18, 374–381. [Google Scholar] [CrossRef]
- D'Amico, S.; Claverie, P.; Collins, T.; Georlette, D.; Gratia, E.; Hoyoux, A.; Meuwis, M.A.; Feller, G.; Gerday, C. Molecular basis of cold adaptation. Philos Trans. R Soc. Lond B Biol. Sci 2002, 357, 917–925. [Google Scholar] [CrossRef]
- Feller, G.; Gerday, C. Psychrophilic enzymes: Hot topics in cold adaptation. Nat. Rev. Microbiol. 2003, 1, 200–208. [Google Scholar] [CrossRef]
- Marx, J.C.; Collins, T.; D'Amico, S.; Feller, G.; Gerday, C. Cold-adapted enzymes from marine antarctic microorganisms. Mar. Biotechnol. 2007, 9, 293–304. [Google Scholar] [CrossRef]
- Collins, T.; De Vos, D.; Hoyoux, A.; Savvides, S.N.; Gerday, C.; Van Beeumen, J.; Feller, G. Study of the active site residues of a glycoside hydrolase family 8 xylanase. J. Mol. Biol. 2005, 354, 425–435. [Google Scholar] [CrossRef]
- Collins, T.; Hoyoux, A.; Dutron, A.; Georis, J.; Genot, B.; Dauvrin, T.; Arnaut, F.; Gerday, C.; Feller, G. Use of glycoside hydrolase family 8 xylanases in baking. J. Cereal Sci. 2006, 43, 79–84. [Google Scholar] [CrossRef]
- Collins, T.; Meuwis, M.A.; Gerday, C.; Feller, G. Activity, stability and flexibility in glycosidases adapted to extreme thermal environments. J. Mol. Biol. 2003, 328, 419–428. [Google Scholar] [CrossRef]
- Garsoux, G.; Lamotte, J.; Gerday, C.; Feller, G. Kinetic and structural optimization to catalysis at low temperatures in a psychrophilic cellulase from the antarctic bacterium pseudoalteromonas haloplanktis. Biochem. J. 2004, 384, 247–253. [Google Scholar] [CrossRef]
- Feller, G.; d'Amico, D.; Gerday, C. Thermodynamic stability of a cold-active alpha-amylase from the antarctic bacterium alteromonas haloplanctis. Biochemistry 1999, 38, 4613–4619. [Google Scholar] [CrossRef]
- Siddiqui, K.S.; Feller, G.; D'Amico, S.; Gerday, C.; Giaquinto, L.; Cavicchioli, R. The active site is the least stable structure in the unfolding pathway of a multidomain cold-adapted alpha-amylase. J. Bacteriol. 2005, 187, 6197–6205. [Google Scholar] [CrossRef]
- Siddiqui, K.S.; Poljak, A.; Guilhaus, M.; De Francisci, D.; Curmi, P.M.; Feller, G.; D'Amico, S.; Gerday, C.; Uversky, V.N.; Cavicchioli, R. Role of lysine versus arginine in enzyme cold-adaptation: Modifying lysine to homo-arginine stabilizes the cold-adapted alpha-amylase from pseudoalteramonas haloplanktis. Proteins 2006, 64, 486–501. [Google Scholar] [CrossRef]
- Aurilia, V.; Parracino, A.; Saviano, M.; Rossi, M.; D'Auria, S. The psychrophilic bacterium pseudoalteromonas halosplanktis tac125 possesses a gene coding for a cold-adapted feruloyl esterase activity that shares homology with esterase enzymes from gamma-proteobacteria and yeast. Gene 2007, 397, 51–57. [Google Scholar] [CrossRef]
- de Pascale, D.; Cusano, A.M.; Autore, F.; Parrilli, E.; di Prisco, G.; Marino, G.; Tutino, M.L. The cold-active lip1 lipase from the antarctic bacterium pseudoalteromonas haloplanktis tac125 is a member of a new bacterial lipolytic enzyme family. Extremophiles 2008, 12, 311–323. [Google Scholar] [CrossRef]
- Aghajari, N.; Van Petegem, F.; Villeret, V.; Chessa, J.P.; Gerday, C.; Haser, R.; Van Beeumen, J. Crystal structures of a psychrophilic metalloprotease reveal new insights into catalysis by cold-adapted proteases. Proteins 2003, 50, 636–647. [Google Scholar] [CrossRef]
- Handelsman, J.; Rondon, M.R.; Brady, S.F.; Clardy, J.; Goodman, R.M. Molecular biological access to the chemistry of unknown soil microbes: A new frontier for natural products. Chem. Biol. 1998, 5, R245–R249. [Google Scholar] [CrossRef]
- Rondon, M.R.; August, P.R.; Bettermann, A.D.; Brady, S.F.; Grossman, T.H.; Liles, M.R.; Loiacono, K.A.; Lynch, B.A.; MacNeil, I.A.; Minor, C.; et al. Cloning the soil metagenome: A strategy for accessing the genetic and functional diversity of uncultured microorganisms. Appl. Environ. Microbiol. 2000, 66, 2541–2547. [Google Scholar]
- Heath, C.; Hu, X.P.; Cary, S.C.; Cowan, D. Identification of a novel alkaliphilic esterase active at low temperatures by screening a metagenomic library from antarctic desert soil. Appl. Environ. Microbiol. 2009, 75, 4657–4659. [Google Scholar] [CrossRef]
- Kim, E.Y.; Oh, K.H.; Lee, M.H.; Kang, C.H.; Oh, T.K.; Yoon, J.H. Novel cold-adapted alkaline lipase from an intertidal flat metagenome and proposal for a new family of bacterial lipases. Appl. Environ. Microbiol. 2009, 75, 257–260. [Google Scholar] [CrossRef]
- Berlemont, R.; Pipers, D.; Delsaute, M.; Angiono, F.; Feller, G.; Galleni, M.; Power, P. Exploring the antarctic soil metagenome as a source of novel cold-adapted enzymes and genetic mobile elements. Revista Argentina de microbiologia 2011, 43, 94–103. [Google Scholar]
- Berlemont, R.; Delsaute, M.; Pipers, D.; D'Amico, S.; Feller, G.; Galleni, M.; Power, P. Insights into bacterial cellulose biosynthesis by functional metagenomics on antarctic soil samples. ISME J. 2009, 3, 1070–1081. [Google Scholar] [CrossRef]
- Arpigny, J.L.; Jaeger, K.E. Bacterial lipolytic enzymes: Classification and properties. Biochem. J. 1999, 343, 177–183. [Google Scholar] [CrossRef]
- Punta, M.; Coggill, P.C.; Eberhardt, R.Y.; Mistry, J.; Tate, J.; Boursnell, C.; Pang, N.; Forslund, K.; Ceric, G.; Clements, J.; et al. The pfam protein families database. Nucleic Acids Res. 2012, 40, D290–D301. [Google Scholar] [CrossRef]
- Marchot, P.; Chatonnet, A. Enzymatic activity and protein interactions in alpha/beta hydrolase fold proteins: Moonlighting versus promiscuity. Protein Pept. Lett. 2012, 19, 132–143. [Google Scholar] [CrossRef]
- Retief, J.D. Phylogenetic analysis using phylip. Meth. in Mol. Biol. 2000, 132, 243–258. [Google Scholar]
- Cheeseman, J.D.; Tocilj, A.; Park, S.; Schrag, J.D.; Kazlauskas, R.J. Structure of an aryl esterase from pseudomonas fluorescens. Acta. Crystallogr. D Biol. Crystallogr. 2004, 60, 1237–1243. [Google Scholar] [CrossRef]
- Otero, C.; Fernández-Pérez, M.; Hermoso, J.A.; Ripoll, M.M. Activation in the family of candida rugosa isolipases by polyethylene glycol. J. Mol. Catal. B Enzym. 2005, 32, 225–229. [Google Scholar] [CrossRef]
- Chu, X.; He, H.; Guo, C.; Sun, B. Identification of two novel esterases from a marine metagenomic library derived from south china sea. Appl. Microbiol. Biotechnol. 2008, 80, 615–625. [Google Scholar] [CrossRef]
- Hong, K.S.; Lim, H.K.; Chung, E.J.; Park, E.J.; Lee, M.H.; Kim, J.C.; Choi, G.J.; Cho, K.Y.; Lee, S.W. Selection and characterization of forest soil metagenome genes encoding lipolytic enzymes. J. Microbiol. Biotechnol. 2007, 17, 1655–1660. [Google Scholar]
- Wei, P.; Bai, L.; Song, W.; Hao, G. Characterization of two soil metagenome-derived lipases with high specificity for p-nitrophenyl palmitate. Arch. Microbiol. 2009, 191, 233–240. [Google Scholar] [CrossRef]
- Khalameyzer, V.; Fischer, I.; Bornscheuer, U.T.; Altenbuchner, J. Screening, nucleotide sequence, and biochemical characterization of an esterase from pseudomonas fluorescens with high activity towards lactones. Appl. Environ. Microbiol. 1999, 65, 477–482. [Google Scholar]
- Itoh, N.; Kawanami, T.; Liu, J.Q.; Dairi, T.; Miyakoshi, M.; Nitta, C.; Kimoto, Y. Cloning and biochemical characterization of co(2+)-activated bromoperoxidase-esterase (perhydrolase) from pseudomonas putida if-3 strain. Biochim. Biophys. Acta 2001, 1545, 53–66. [Google Scholar] [CrossRef]
- Pelletier, I.; Altenbuchner, J. A bacterial esterase is homologous with non-haem haloperoxidases and displays brominating activity. Microbiology 1995, 141, 459–468. [Google Scholar] [CrossRef]
- Loo, T.L.; Burger, J.W.; Adamson, R.H. Bromination of phthalein dyes by the uterus of the dogfish, squalus acanthias. Proc. Soc. Exp. Biol. Med. 1963, 114, 60–63. [Google Scholar]
- Polgar, L.; Szigetvari, A.; Low, M.; Korodi, I.; Balla, E. Metalloendopeptidase qg. Isolation from escherichia coli and characterization. Biochem. J. 1991, 273, 725–731. [Google Scholar]
- Wang, G.; Meng, K.; Luo, H.; Wang, Y.; Huang, H.; Shi, P.; Pan, X.; Yang, P.; Yao, B. Molecular cloning and characterization of a novel sgnh arylesterase from the goat rumen contents. Appl. Microbiol. Biotechnol. 2011, 91, 1561–1570. [Google Scholar] [CrossRef]
- Park, S.; Morley, K.L.; Horsman, G.P.; Holmquist, M.; Hult, K.; Kazlauskas, R.J. Focusing mutations into the p. Fluorescens esterase binding site increases enantioselectivity more effectively than distant mutations. Chem. Biol. 2005, 12, 45–54. [Google Scholar] [CrossRef]
- Fenster, K.M.; Parkin, K.L.; Steele, J.L. Nucleotide sequencing, purification, and biochemical properties of an arylesterase from lactobacillus casei lila. J. Dairy Sci. 2003, 86, 2547–2557. [Google Scholar] [CrossRef]
- Liu, A.M.F.; Somers, N.A.; Kazlauskas, R.J.; Brush, T.S.; Zocher, F.; Enzelberger, M.M.; Bornscheuer, U.T.; Horsman, G.P.; Mezzetti, A.; Schmidt-Dannert, C.; et al. Mapping the substrate selectivity of new hydrolases using colorimetric screening: Lipases from bacillus thermocatenulatus and ophiostoma piliferum, esterases from pseudomonas fluorescens and streptomyces diastatochromogenes. Tetrahedron Asymmetry 2001, 12, 545–556. [Google Scholar] [CrossRef]
- Gerday, C.; Aittaleb, M.; Arpigny, J.L.; Baise, E.; Chessa, J.P.; Garsoux, G.; Petrescu, I.; Feller, G. Psychrophilic enzymes: A thermodynamic challenge. Biochimica et biophysica acta 1997, 1342, 119–131. [Google Scholar] [CrossRef]
- Sonan, G.K.; Receveur-Brechot, V.; Duez, C.; Aghajari, N.; Czjzek, M.; Haser, R.; Gerday, C. The linker region plays a key role in the adaptation to cold of the cellulase from an antarctic bacterium. Biochem. J. 2007, 407, 293–302. [Google Scholar] [CrossRef]
- Bauvois, C.; Jacquamet, L.; Huston, A.L.; Borel, F.; Feller, G.; Ferrer, J.L. Crystal structure of the cold-active aminopeptidase from colwellia psychrerythraea, a close structural homologue of the human bifunctional leukotriene a4 hydrolase. J. Biol. Chem. 2008, 283, 23315–23325. [Google Scholar]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Berlemont, R.; Jacquin, O.; Delsaute, M.; La Salla, M.; Georis, J.; Verté, F.; Galleni, M.; Power, P. Novel Cold-Adapted Esterase MHlip from an Antarctic Soil Metagenome. Biology 2013, 2, 177-188. https://doi.org/10.3390/biology2010177
Berlemont R, Jacquin O, Delsaute M, La Salla M, Georis J, Verté F, Galleni M, Power P. Novel Cold-Adapted Esterase MHlip from an Antarctic Soil Metagenome. Biology. 2013; 2(1):177-188. https://doi.org/10.3390/biology2010177
Chicago/Turabian StyleBerlemont, Renaud, Olivier Jacquin, Maud Delsaute, Marcello La Salla, Jacques Georis, Fabienne Verté, Moreno Galleni, and Pablo Power. 2013. "Novel Cold-Adapted Esterase MHlip from an Antarctic Soil Metagenome" Biology 2, no. 1: 177-188. https://doi.org/10.3390/biology2010177
APA StyleBerlemont, R., Jacquin, O., Delsaute, M., La Salla, M., Georis, J., Verté, F., Galleni, M., & Power, P. (2013). Novel Cold-Adapted Esterase MHlip from an Antarctic Soil Metagenome. Biology, 2(1), 177-188. https://doi.org/10.3390/biology2010177