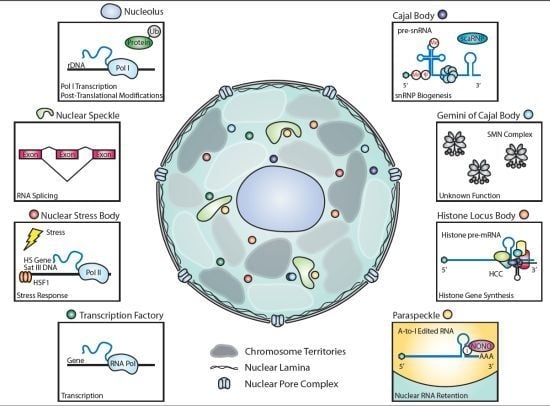
The Role of Nuclear Bodies in Gene Expression and Disease
Abstract
:1. Introduction

2. Nuclear Bodies and Mechanisms of Gene Expression
2.1. Nucleolus
2.1.1. Discovery
2.1.2. Key Components
2.1.3. Functions of Key Components
| Nuclear Body Component | Synonyms | Notes | Nuclear Body Component Localization References |
|---|---|---|---|
| Nucleolus | |||
| Proteins | |||
| Nucleophosmin | B23 | Multifunctional chaperone | [19] |
| Nucleolin | C23 | pre-rRNA processing RNA polymerase I transcription | [19] |
| Fibrillarin | Box C/D snoRNP component pre-rRNA 2'-O-methyltransferase | [20] | |
| NHP2L1 | 15.5K NHPX | Box C/D snoRNP component pre-rRNA 2'-O-methylation | [21] |
| NOP56 | Box C/D snoRNP component pre-rRNA 2'-O-methylation | [22] | |
| NOP58 | Box C/D snoRNP component pre-rRNA 2'-O-methylation | [22] | |
| Dyskerin | DKC1 NAP57 NOLA4 | H/ACA snoRNP component pre-rRNA pseudouridylase Telomerase component | [23] |
| GAR1 | NOLA1 | H/ACA snoRNP component pre-rRNA pseudouridylation Telomerase component | [24] |
| NHP2 | NOLA2 | H/ACA snoRNP component pre-rRNA pseudouridylation Telomerase component | [25] |
| NOP10 | NOLA3 | H/ACA snoRNP component pre-rRNA pseudouridylation Telomerase component | [25] |
| NOPP140 | NOLC1 | Box C/D snoRNP chaperone Transcription | [26] |
| RNA polymerase I | Transcription | [12,13,27] | |
| 40S ribosomal subunit | 40S small ribosomal subunit Translation | [12,13] | |
| 60S ribosomal subunit | 60S large ribosomal subunit Translation | [12,13] | |
| Nucleolus | |||
| RNAs | |||
| U3 snoRNA | Box C/D snoRNA pre-rRNA processing | [28] | |
| U8 snoRNA | Box C/D snoRNA pre-rRNA processing | [28,29] | |
| U13 snoRNA | Box C/D snoRNA pre-rRNA processing | [28] | |
| U14 snoRNA | Box C/D snoRNA pre-rRNA processing | [29] | |
| U17 snoRNA | E1 snoRNA | Box H/ACA snoRNA pre-rRNA processing | [30] |
| E2 snoRNA | Box H/ACA snoRNA pre-rRNA processing | [30] | |
| E3 snoRNA | Box H/ACA snoRNA pre-rRNA processing | [30] | |
| Nuclear Speckle | |||
| Proteins | |||
| SRSF1 | SF2 ASF SRp30a | SRSF family Constitutive and alternative splicing | [31] |
| SRSF2 | SC35 SRp30b | SRSF family Constitutive and alternative splicing | [32] |
| SRSF3 | SRp20 | SRSF family Constitutive and alternative splicing | [31] |
| SRSF4 | SRp75 | SRSF family Constitutive and alternative splicing | [33,34] |
| SRSF5 | SRp40 | SRSF family Constitutive and alternative splicing | [34] |
| SRSF6 | SRp55 | SRSF family Constitutive and alternative splicing | [33,34] |
| SRSF7 | 9G8 | SRSF family Constitutive and alternative splicing | [34] |
| CLK | STY | Dual specificity protein kinase SRSF phosphorylation | [35] |
| PRP3 | U4/U6 snRNP component pre-mRNA splicing | [34] | |
| PRP6 | U4/U6-U5 tri-snRNP component pre-mRNA splicing | [34] | |
| PRP8 | U5 snRNP component pre-mRNA splicing | [34] | |
| U5 snRNP 200 kDa protein | BRR2 | U5 snRNP component pre-mRNA splicing | [34] |
| U5 snRNP 116 kDa break/> protein | SNU114 | U5 snRNP component pre-mRNA splicing | [33,34] |
| CPSF160 | CPSF1 | CPSF complex pre-mRNA 3' end processing | [34] |
| CPSF100 | CPSF2 | CPSF complex pre-mRNA 3' end processing | [34] |
| CPSF73 | CPSF3 | CPSF complex pre-mRNA 3' end processing | [34] |
| CPSF30 | CPSF4 | CPSF complex pre-mRNA 3' end processing | [34] |
| CFIm68 | CPSF6 | CFIm complex pre-mRNA 3' end processing | [34] |
| CSTF64 | CSTF2 | CSTF complex pre-mRNA 3' end processing | [34] |
| RNA polymerase II | Transcription | [34,36] | |
| SON | pre-mRNA splicing | [34] | |
| MAGOH | Exon junction complex mRNA export and NMD | [34] | |
| eIF4AIII | DDX48 | Exon junction complex mRNA export and NMD | [34] |
| RNPS1 | Exon junction complex mRNA export and NMD | [33,34] | |
| Y14 | RBM8A | Exon junction complex mRNA export and NMD | [34] |
| Aly/REF | Exon junction complex mRNA export and NMD | [34] | |
| RAE1 | mRNA export | [34] | |
| RNAs | |||
| U1 snRNA | U1 snRNP component pre-mRNA splicing | [37] | |
| U2 snRNA | U2 snRNP component pre-mRNA splicing | [37] | |
| MALAT1 RNA | NEAT2 | Long ncRNA | [38] |
| Poly(A)+ RNA | RNA with a poly(A) tail | [39] | |
| Nuclear Stress Body | |||
| Proteins | |||
| HSF1 | Heat shock transcription factor family Transcription | [40] | |
| HSF2 | Heat shock transcription factor family Transcription | [41] | |
| SAF-B | HAP HET | S/MAR binding protein Transcription | [42] |
| Sam68 | KHDRBS1 | Transcription Alternative splicing | [43] |
| SRSF1 | ASF SF2 | SRSF family Constitutive and alternative splicing | [43] |
| SRSF7 | 9G8 | SRSF family Constitutive and alternative splicing | [43] |
| SRSF9 | SRp30c | SRSF family Constitutive and alternative splicing | [43] |
| RNA polymerase II | Transcription | [44] | |
| RNA | |||
| Satellite III ncRNA | Long ncRNA transcribed from the pericentric heterochromatic 9p12 locus | [44] | |
| Transcription Factory | |||
| Proteins | |||
| RNA polymerase I | Transcription | [45,46] | |
| RNA polymerase II | Transcription | [45,46] | |
| RNA polymerase III | Transcription | [45,46] | |
| Cajal Body | |||
| Proteins | |||
| Coilin | Function unknown | [47,48] | |
| Fibrillarin | Box C/D snoRNP component pre-rRNA 2'-O-methyltransferase | [49] | |
| NHP2L1 | 15.5K NHPX | Box C/D snoRNP component pre-rRNA 2'-O-methylation | [21,50] |
| NOP56 | Box C/D snoRNP component pre-rRNA 2'-O-methylation | [50] | |
| NOP58 | Box C/D snoRNP component pre-rRNA 2'-O-methylation | [50] | |
| Dyskerin | DKC1 NAP57 NOLA4 | H/ACA snoRNP component pre-rRNA pseudouridylase Telomerase component | [51] |
| GAR1 | NOLA1 | H/ACA snoRNP component pre-rRNA pseudouridylation Telomerase component | [52] |
| NHP2 | NOLA2 | H/ACA snoRNP component pre-rRNA pseudouridylation Telomerase component | [52] |
| NOP10 | NOLA3 | H/ACA snoRNP component pre-rRNA pseudouridylation Telomerase component | [52] |
| TERT | Telomerase component Reverse transcriptase | [53] | |
| TCAB1 | WRAP53 | Telomerase component | [54] |
| NOPP140 | NOLC1 | Box C/D snoRNP chaperone Transcription | [18] |
| SMN1 | GEMIN1 | SMN complex Spliceosomal snRNP biogenesis | [55] |
| RNAs | |||
| U85 scaRNA | scaRNA snRNA modification | [56] | |
| U87 scaRNA | scaRNA snRNA modification | [56] | |
| U88 scaRNA | scaRNA snRNA modification | [56] | |
| U89 scaRNA | scaRNA snRNA modification | [56] | |
| U90 scaRNA | scaRNA snRNA modification | [56] | |
| U91 scaRNA | scaRNA snRNA modification | [56] | |
| U92 scaRNA | scaRNA snRNA modification | [56] | |
| U2 snRNA | U2 snRNP component pre-mRNA splicing | [57,58] | |
| U4 snRNA | U4 snRNP component pre-mRNA splicing | [57,58] | |
| U5 snRNA | U5 snRNP component pre-mRNA splicing | [57,58] | |
| U6 snRNA | U6 snRNP component pre-mRNA splicing | [57,58] | |
| U3 snoRNA | Box C/D snoRNA pre-rRNA processing | [59] | |
| U8 snoRNA | Box C/D snoRNA pre-rRNA processing | [59] | |
| U14 snoRNA | Box C/D snoRNA pre-rRNA processing | [59] | |
| TERC RNA | TR | Telomerase complex | [53] |
| Gemini of Cajal Body | |||
| Proteins | |||
| SMN1 | GEMIN1 | SMN complex Spliceosomal snRNP biogenesis | [60] |
| GEMIN2 | SIP1 | SMN complex Spliceosomal snRNP biogenesis | [61] |
| GEMIN3 | SMN complex Spliceosomal snRNP biogenesis | [62] | |
| GEMIN4 | SMN complex Spliceosomal snRNP biogenesis | [63] | |
| GEMIN5 | SMN complex Spliceosomal snRNP biogenesis | [64] | |
| GEMIN6 | SIP2 | SMN complex Spliceosomal snRNP biogenesis | [65] |
| GEMIN7 | SIP3 | SMN complex Spliceosomal snRNP biogenesis | [66] |
| GEMIN8 | SMN complex Spliceosomal snRNP biogenesis | [67] | |
| ZPR1 | SMN complex SMN localization | [68] | |
| Histone Locus Body | |||
| Proteins | |||
| NPAT | p220 | Histone gene transcription | [69] |
| SLBP | HBP | Histone pre-mRNA 3' end processing | [70] |
| LSm10 | U7 snRNP component Histone pre-mRNA 3' end processing | [70] | |
| LSm11 | U7 snRNP component Histone pre-mRNA 3' end processing | [70,71] | |
| FLASH | CASP8AP2 | Histone gene transcription | [72] |
| NELF A | WHSC2 | NELF complex Histone pre-mRNA 3' end processing | [73] |
| NELF B | COBRA1 | NELF complex Histone pre-mRNA 3' end processing | [73] |
| NELF C/D | TH1L | NELF complex Histone pre-mRNA 3' end processing | [73] |
| NELF E | RDBP | NELF complex Histone pre-mRNA 3' end processing | [73] |
| HiNF-P | Transcription | [74] | |
| ZPR1 | NPAT, SMN, coilin localization to HLBs | [75] | |
| Coilin | Function unknown | [76] | |
| RNA | |||
| U7 snRNA | U7 snRNP component Histone pre-mRNA 3' end processing | [71] | |
| DNA | |||
| Histone loci | Replication-dependent histone genes | [71,77] | |
| Paraspeckle | |||
| Proteins | |||
| NONO | p54nrb | DBHS family | [78] |
| PSP1 | PSPC1 | DBHS family | [78] |
| PSP2 | CoAA RBM14 | Transcription Splicing | [78] |
| SFPQ | PSF | DBHS family | [79] |
| CFIm68 | CPSF6 | CFIm complex pre-mRNA 3' end processing | [80] |
| RNAs | |||
| NEAT1 RNA | MEN ε/β VINC | Mammal-specific long ncRNA | [81,82,83] |
| Ctn RNA | Mouse-specific isoform transcribed from the Slc7a2 locus | [79] | |
| Nuclear Body Component | Model System | Experimental Manipulation | Phenotype | References |
|---|---|---|---|---|
| Nucleolus | ||||
| Proteins | ||||
| ? | African clawed frog | Anucleolate mutant | Absence of rRNA synthesis Developmental arrest | [8] |
| Nucleophosmin | HeLa cells | Transduction of NPM shuttling defective mutants | ⬇ Nuclear export of 40S and 60S ribosomal subunits ⬇ Protein synthesis | [14] |
| Nucleolin | HeLa cells | siRNA KD of NCL | ⬇ RNA Pol I transcription | [16] |
| Fibrillarin | Budding yeast | Conditionally lethal allele of NOP1 | ⬇ 18S rRNA ⬇ Ribosome biogenesis ⬇ Growth rate | [84] |
| Dyskerin | HeLa cells | siRNA KD of DKC1 | ⬇ Pseudouridylation of rRNAs ⬇ Ribosome affinity for substrate ⬇ Translational fidelity | [85] |
| Dyskerin | Cultured mouse embryonic fibroblasts | Hypomorphic allele of Dkc1 | ⬇ Pseudouridylation of rRNAs ⬇ Ribosome affinity for substrate ⬇ Translational fidelity | [85] |
| Dyskerin | Budding yeast | Hypomorphic allele of CBF5 | ⬇ Pseudouridylation of rRNAs ⬇ Ribosome affinity for substrate ⬇ Translational fidelity | [85] |
| RNA Pol I | Human peripheral blood lymphoblasts and cultured fibroblasts | Actinomycin Dtreatment | Condensation of nucleolar chromatin Coilin enriched in nucleolar caps | [86] |
| RPS19 | DBA patient-derived CD34- cells from bone marrow | None | ⬇Mature 40S ribosomal subunit pre-rRNA processing defect | [87] |
| RPS19 | Human erythroleukemia cells | siRNA KD of RPS19 | pre-rRNA processing defect | [87] |
| RNA | ||||
| U3 snoRNA | Budding yeast | Conditionally repressible SNR17A gene | ⬇ 18S rRNA ⬇ Growth rate | [88] |
| Nuclear Speckle | ||||
| Proteins | ||||
| SRSF1 | HeLa cells | siRNA KD of SRSF1 | ⬇ Nuclear speckle localization of pre-mRNA processing factors Enlarged nuclear speckles | [89] |
| SRSF1 | Human osteosarcoma cells | siRNA KD of SRSF1 | ⬇ Transcription | [89] |
| SRSF2 | Human osteosarcoma cells | siRNA KD of SRSF2 | ⬇ Transcription | [89] |
| CLK | Human epidermoid carcinoma cells | Overexpression of murine CLK | ⬇ pre-mRNA splicing Disrupted nuclear speckles | [90] |
| CLK | Human epidermoid carcinoma cells | Overexpression of catalytically inactive murine CLK | Retention of hypophosphorylated SRSF proteins in nuclear speckles | [90] |
| PRPF6 | HeLa cells | siRNA KD of PRPF6 | ⬇ MALAT1 RNA ⬇ Localization of MALAT1 to nuclear speckles Nuclear speckles not disrupted | [91] |
| SON | HeLa cells | siRNA KD of SON | ⬇ Localization of MALAT1 to nuclear speckles | [91] |
| SON | HeLa cells | siRNA KD of SON | ⬇ Localization of pre-mRNA processing factors to nuclear speckles ⬇ Localization of core EJC proteins to nuclear specklesCell cycle arrest | [92] |
| Aly/REF | HeLa cells | siRNA KD of ALYREF | ⬇ mRNA export | [93] |
| Aly/REF | HeLa cells | siRNA KD of ALYREF | ⬇ mRNA export ⬇ Poly(A)+ RNA in nuclear speckles | [94] |
| RNAs | ||||
| MALAT1 | HeLa cells | ASO KD of MALAT1 | ⬇ Nuclear speckle integrityAberrant alternative splicing ⬇ SRSF1 and SRSF2 Altered ratio of dephosphorylated to phosphorylated SRSF proteins | [91] |
| Malat1 | Mouse | KO | Localization of nuclear speckle proteins unaffected Viable and fertile | [95,96] |
| U1 snRNA | HeLa cells | ASO KD of U1 snRNA | Splicing inhibition Enlarged speckles ⬇ Number of nuclear speckles ⬇ Transcription | [97] |
| U2 snRNA | HeLa cells | ASO KD of U2 snRNA | Splicing inhibition Enlarged speckles ⬇ Number of nuclear speckles ⬇ Transcription | [97] |
| Nuclear Stress Body | ||||
| Proteins | ||||
| HSF1 | Cultured human embryonic kidney cells | shRNA KD of HSF1 | ⬇ Satellite III transcription | [98] |
| HSF1 | HeLa cells | shRNA KD of HSF1 | ⬇ HSF2 protein No nSBs upon heat shock | [98] |
| HSF2 | Cultured human embryonic kidney cells | siRNA KD of HSF2 | ⬇ Satellite III transcription No effect on HSF1 localization to nSBs upon heat shock | [98] |
| Transcription Factory | ||||
| Proteins | ||||
| RNA Pol I | Human cells | Actinomycin D treatment | Coilin enriched in nucleolar caps | [86] |
| RNA Pol II | Human cells | α-Amanitin or DRB treatment | Chromatin decondensation Loss of nucleolar structure | [86] |
| RNA Pol II | HeLa cells | α-Amanitin treatment | ⬇ BrUTP incorporation | [99,100] |
| RNA Pol II | Canine kidney cells | α-Amanitin, DRB, H8, or actinomycin D treatment | Redistribution of RNA Pol II to enlarged nuclear speckles | [36] |
| RNA Pol III | HeLa cells | α-Amanitin treatment | #x2B07; BrUTP incorporation | [100] |
| Cajal Body | ||||
| Proteins | ||||
| Coilin | Mouse | KO | ⬇ Viability Residual CBs present No SMN localization to CBs No Sm protein localization to CBs | [101,102] |
| Coilin | African clawed frog oocyte | Coilin removal by IP | CBs present No Sm protein localization to CBs | [103] |
| Coilin | Fruit fly | Amorphic alleles of coil | No CBs No effect on HLBs Viable and fertile | [76] |
| Coilin | Fruit fly | Amorphic alleles of coil | No CBs No scaRNA localization to CBs snRNAs properly modified | [104] |
| Dyskerin | DKC patient-derived iPSCs | None | ⬇ Telomerase assembly ⬇ Telomere synthesis | [105] |
| TERT | DKC patient-derived iPSCs | None | ⬇ Telomerase levels ⬇ Telomere synthesis | [105] |
| TCAB1 | DKC patient-derived iPSCs | None | Telomerase activity unaffected ⬇ Telomerase localization to CBs ⬇ Telomere synthesis | [105] |
| TCAB1 | HeLa cells | shRNA KD of WRAP53 | ⬇ TERC localization to CBs ⬇ TERC localization to telomeres ⬇ Telomere synthesis | [54] |
| NOPP140 | Cultured SMA patient-derived fibroblasts | None | ⬇ Localization of dyskerin and GAR1 to CBs ⬇ Localization of NOPP140 correlated with disease severity | [106] |
| SMN1 | Cultured SMA patient-derived lymphoblasts and fibroblasts | None | ⬇ Gems ⬇ SMN protein | [107,108] |
| SMN1 | HeLa cells | siRNA KD of SMN1 | Defects in CB formation snRNPs absent in coilin foci | [109] |
| SMN1 | HeLa cells | siRNA KD of SMN1 | ⬇ Gem-associated proteins ⬇ snRNP assembly Gems absent | [110,111] |
| SMN1 | Mouse | Neuron-specific exon 7 deletion of Smn1 | Coilin aggregates Gems absent SMA phenotype | [112] |
| SMN1 | Mouse | SMA models varying in disease severity | ⬇ SMN complex proteins ⬇ snRNP biogenesis Correlation with disease severity | [113] |
| SMN1 | Mouse | Human SMN2 and SMN∆7 expression in Smn1-/- background | ⬇ SMN complex proteins ⬇ snRNP biogenesis Tissue-specific snRNA alterations Widespread splicing defects SMA phenotype | [114] |
| SMN1 | Fission yeast | Temperature-sensitive degron allele of smn1 | Differential snRNP biogenesis Splicing defects | [115] |
| Gemini of Cajal Body | ||||
| Proteins | ||||
| SMN1 | Cultured SMA patient-derived lymphoblasts and fibroblasts | None | ⬇ Gems ⬇ SMN protein | [107,108] |
| SMN1 | HeLa cells | siRNA KD of SMN1 | Defects in CB formation snRNPs absent in coilin foci | [109] |
| SMN1 | HeLa cells | siRNA KD of SMN1 | ⬇ Gem-associated proteins ⬇ snRNP assembly Gems absent | [110,111] |
| SMN1 | Mouse | Neuron-specific exon 7 deletion of Smn1 | Coilin aggregates Gems absent SMA phenotype | [112] |
| SMN1 | Mouse | SMA models varying in disease severity | ⬇ SMN complex proteins ⬇ snRNP biogenesis Correlation with disease severity | [113] |
| SMN1 | Mouse | Human SMN2 and SMN∆7 expression in Smn1-/- background | ⬇ SMN complex proteins ⬇ snRNP biogenesis Tissue-specific snRNA alterations Widespread splicing defects SMA phenotype | [114] |
| SMN1 | Fission yeast | Temperature-sensitive degron allele of smn1 | Differential snRNP biogenesis Splicing defects | [115] |
| GEMIN2 | HeLa cells | siRNA KD of GEMIN2 | ⬇ GEMIN3 protein ⬇ snRNP biogenesis | [110,111] |
| GEMIN3 | HeLa cells | siRNA KD of GEMIN3 | ⬇ GEMIN4 protein ⬇ snRNP biogenesis | [110,111] |
| GEMIN3 | Fruit fly | LOF alleles of Gem3 | ⬇ SMN protein Motor defects Larval lethal | [116] |
| GEMIN4 | HeLa cells | siRNA KD of GEMIN4 | ⬇ GEMIN3 protein ⬇ snRNP biogenesis | [110,111] |
| GEMIN5 | HeLa cells | siRNA KD of GEMIN5 | No effect on gems No effect on snRNP biogenesis | [110,111] |
| GEMIN6 | HeLa cells | siRNA KD of GEMIN6 | No effect on gems ⬇ snRNP biogenesis | [110,111] |
| GEMIN7 | HeLa cells | siRNA KD of GEMIN7 | ⬇ snRNP biogenesis | [111] |
| ZPR1 | HeLa cells | ASO KD of ZPR1 | ⬇ SMN localization to gems and CBs | [68] |
| ZPR1 | Mouse | KO | ⬇ Localization of SMN and coilin to gems and CBs Mislocalization of snRNPs Embryonic lethal | [117] |
| ZPR1 | Mouse | Heterozygous KO | Mislocalization of SMN Motor neuron degeneration Motor defects | [118] |
| ZPR1 | Cultured mouse motor neuron-like cells | siRNA KD of Zpr1 | ⬇ Localization of SMN to gems and CBs Mislocalization of snRNPs Axonal defects | [117] |
| Histone Locus Body | ||||
| Proteins | ||||
| NPAT | Mouse | Retroviral insertion of Npat | Embryonic lethal | [119] |
| SLBP | Human osteosarcoma cells | siRNA KD of SLBP | Aberrant histone RNA processing S phase block ⬇ Cell proliferation ⬇ Histone mRNA and protein | [120] |
| SLBP | HeLa cells | shRNA KD of SLBP | Aberrant histone RNA processing | [73] |
| SLBP | Fruit fly | LOF alleles of Slbp | Aberrant histone pre-mRNA processing Female sterile Lethal | [121] |
| SLBP | Fruit fly | LOF allele of Slbp | No effect on HLBs | [122] |
| LSm10 | Fruit fly | LOF alleles of Lsm10 | Aberrant histone pre-mRNA processing No U7 snRNA localization to HLBs Lethal | [123] |
| LSm11 | Fruit fly | LOF alleles of Lsm11 | ⬇ Lsm10 protein Aberrant histone pre-mRNA processing No U7 snRNA localization to HLBs Lethal | [123] |
| FLASH | HeLa cells | shRNA KD of CASP8AP2 | ⬇ Histone mRNA ⬇ Histone protein S phase block | [124] |
| FLASH | Human breast cancer cells | shRNA KD of CASP8AP2 | No HLBs | [124] |
| FLASH | Mouse | KO | Embryonic lethal | [125] |
| NELF E | HeLa cells | shRNA KD of RDBP | ⬇ NELF complex proteins ⬇ Cell proliferation Aberrant histone RNA processing | [73] |
| HiNF-P | Human glioblastoma cells | ASO and siRNA KD of HINFP | ⬇ Histone 4 gene expression ⬇ RNA Pol II and NPAT histone 4 promoter occupancy | [74] |
| HiNF-P | Mouse | Amorphic allele of Hinfp | ⬇ Histone 4 gene expression Embryonic lethal | [126] |
| ZPR1 | HeLa cells | siRNA and ASO KD of ZPR1 | Mislocalization of SMN, NPAT, coilin and Sm proteins S phase block ⬇ Transcription | [75] |
| ZPR1 | Mouse | KO | Embryonic lethal | [117] |
| Coilin | Human breast cancer cells | shRNA KD of COIL | No effect on HLBs | [124] |
| Coilin | Cultured mouse embryonic fibroblasts | shRNA KD of Coil | No effect on HLBs | [124] |
| Coilin | Fruit fly | Amorphic alleles of coil | No CBs No effect on HLBs Viable and fertile | [76] |
| RNA | ||||
| U7 RNA | Fruit fly | Null alleles of U7 snRNA | Aberrant histone pre-mRNA processing No effect on HLBs No Lsm10 and Lsm11 localization to HLBs Viable, but sterile | [122,123,127] |
| DNA | ||||
| Histone locus | Fruit fly | Histone locus deletion mutant Df(2L)DS6 | Smaller proto-HLBs | [122,128] |
| Paraspeckle | ||||
| Proteins | ||||
| NONO | HeLa cells | siRNA KD of NONO | ⬇ Paraspeckles | [83] |
| NONO | Mouse | Chondrocyte lineage-specific expression of truncated murine NONO | Dwarfism ⬇ Chondrogenesis | [129] |
| NONO | Cultured mouse chondrogenic cells | siRNA KD of Nono | ⬇ Sox9-dependent Col2a1 promoter activity and expression | [129] |
| NONO | Cultured mouse embryonic fibroblasts | siRNA KD of Nono | ⬇ Circadian rhythm | [130] |
| NONO | Fruit fly | Hypomorphic allele of nonA | ⬇ Circadian rhythm | [130] |
| PSP1 | HeLa cells | siRNA KD of PSPC1 | No effect on paraspeckles | [83] |
| SFPQ | HeLa cells | siRNA KD of SFPQ | No paraspeckles | [83] |
| Paraspeckle | ||||
| RNAs | ||||
| NEAT1RNA | HeLa cells | siRNA or ASO KD of NEAT1 | No paraspeckles | [81,82,83,131] |
| NEAT1 RNA | Human osteosarcoma cells | shRNA KD of NEAT1 | No paraspeckles Normal circadian rhythm | [132] |
| Neat1 RNA | Mouse | KO | No paraspeckles Viable and fertile | [133] |
| Ctn RNA | Cultured mouse mammary tumor and macrophage cells | ASO KD of Ctn | No effect on paraspeckles ⬇ Ctn RNA and Cat2 mRNA | [79] |
2.1.4. Mechanisms of Gene Expression and the Nucleolus
2.1.5. Human Diseases Associated with the Nucleolus
| Gene | Human Disease | OMIM No. | Phenotype | Disease Gene Identification References |
|---|---|---|---|---|
| Nucleolus | ||||
| TCOF1 | Treacher Collins syndrome 1 | 154500 | Craniofacial abnormalities | [146] |
| POLR1D | Treacher Collins syndrome 2 | 613717 | Craniofacial abnormalities | [147] |
| POLR1C | Treacher Collins syndrome 3 | 248390 | Craniofacial abnormalities | [147] |
| RPS19 | Diamond-Blackfan anemia 1 | 105650 | Hypoplastic anemia | [148] |
| RPS24 | Diamond-Blackfan anemia 3 | 610629 | Hypoplastic anemia | [149] |
| RPS17 | Diamond-Blackfan anemia 4 | 612527 | Hypoplastic anemia | [150] |
| RPL35A | Diamond-Blackfan anemia 5 | 612528 | Hypoplastic anemia | [151] |
| RPL5 | Diamond-Blackfan anemia 6 | 612561 | Hypoplastic anemia | [152] |
| RPL11 | Diamond-Blackfan anemia 7 | 612562 | Hypoplastic anemia | [152] |
| RPS7 | Diamond-Blackfan anemia 8 | 612563 | Hypoplastic anemia | [152] |
| RPS10 | Diamond-Blackfan anemia 9 | 613308 | Hypoplastic anemia | [153] |
| RPS26 | Diamond-Blackfan anemia 10 | 613309 | Hypoplastic anemia | [153] |
| RPL26 | Diamond-Blackfan anemia 11 | 614900 | Hypoplastic anemia | [154] |
| WRN | Werner syndrome | 277700 | Premature aging syndrome | [155] |
| BLM | Bloom syndrome | 210900 | Growth deficiency Cancer predisposition | [156] |
| Nuclear Speckle | ||||
| PRPF3 | Retinitis pigmentosa 18 | 601414 | Retinal degeneration | [157] |
| PRPF6 | Retinitis pigmentosa 60 | 613983 | Retinal degeneration ⬇ pre-mRNA splicing | [158] |
| PRPF8 | Retinitis pigmentosa 13 | 600059 | Retinal degeneration | [159] |
| SNRNP200 | Retinitis pigmentosa 33 | 610359 | Retinal degeneration ⬇ U4/U6 snRNA unwinding | [160] |
| EFTUD2 | Mandibulofacial dysostosis, Guion-Almeida type | 610536 | Facial dysmorphism Progressive microcephaly Developmental delay Speech delay | [161] |
| RBM8A | Thrombocytopenia-absent radius syndrome | 274000 | Platelet reduction Radial bone aplasia | [162] |
| Transcription Factory | ||||
| POLR1D | Treacher Collins syndrome 2 | 613717 | Craniofacial abnormalities | [147] |
| POLR1C | Treacher Collins syndrome 3 | 248390 | Craniofacial abnormalities | [147] |
| POLR3A | Hypomyelinating leukodystrophy 7 | 607694 | Hypomyelination Motor dysfunction ± Abnormal dentition ± Hypogonadism | [163,164] |
| POLR3B | Hypomyelinating leukodystrophy 8 | 614381 | Hypomyelination Motor dysfunction ± Abnormal dentition ± Hypogonadism | [164,165] |
| MED12 | Lujan-Fryns syndrome | 309520 | Marfanoid habitus Intellectual disability | [166] |
| MED12 | Opitz-Kaveggia syndrome | 305450 | Facial dysmorphism Hypotonia Intellectual disability | [167] |
| MED17 | Postnatal progressive microcephaly with seizures and brain atrophy | 613668 | Progressive microcephaly Brain atrophy Developmental retardation | [168] |
| MED23 | Mental retardation, autosomal recessive 18 | 614249 | Intellectual disability | [169] |
| MED25 | Charcot-Marie-Tooth disease, type 2B2 | 605589 | Distal muscle weakness and atrophy Sensory loss | [170] |
| ERCC2 | Xeroderma pigmentosum, group D | 278730 | Sun sensitivity Increased risk for cancer | [171] |
| ERCC2 | Cerebrooculofacioskeletal syndrome 2 | 610756 | Microcephaly Ocular abnormalities Multiple joint contractures Dysmorphic features | [172] |
| ERCC2 | Trichothiodystrophy | 601675 | Brittle hair and nails Ichthyotic skin Growth delay Intellectual disability | [173] |
| ERCC3 | Xeroderma pigmentosum, group B | 610651 | Sun sensitivity Increased risk for cancer | [174] |
| ERCC3 | Trichothiodystrophy | 601675 | Brittle hair and nails Ichthyotic skin Growth delay Intellectual disability | [175] |
| GTF2H5 | Trichothiodystrophy | 601675 | Brittle hair and nails Ichthyotic skin Growth delay Intellectual disability | [176] |
| Cajal Body | ||||
| SMN1 | Spinal muscular atrophy, type I Spinal muscular atrophy, type II Spinal muscular atrophy, type III Spinal muscular atrophy, type IV | 253300 253550 253400 271150 | Lower motor neuron degeneration ⬇ SMN protein ⬇ Gems | [177] |
| DKC1 | Dyskeratosis congenita, X-linked | 305000 | Nail dystrophy Lacy skin pigmentation Oral leukoplakia ⬇ TERC RNA ⬇ Telomerase assembly ⬇ Telomerase activity | [178] |
| DKC1 | Hoyeraal-Hreidarsson syndrome | 300240 | Growth delay Immunodeficiency Cerebellar hypoplasia | [179] |
| NHP2 | Dyskeratosis congenita, autosomal recessive 2 | 613987 | Nail dystrophy Lacy skin pigmentation Oral leukoplakia ⬇ Telomerase assembly | [180] |
| NOP10 | Dyskeratosis congenita, autosomal recessive 1 | 224230 | Nail dystrophy Lacy skin pigmentation Oral leukoplakia ⬇ Telomerase assembly | [181] |
| TERT | Dyskeratosis congenita, autosomal dominant 2 Dyskeratosis congenita, autosomal recessive 4 | 613989 | Nail dystrophy Lacy skin pigmentation Oral leukoplakia ⬇ Telomerase activity | [182,183] |
| WRAP53 | Dyskeratosis congenita, autosomal recessive 3 | 613988 | Nail dystrophy Lacy skin pigmentation Oral leukoplakia ⬇ Telomerase localization to CBs | [184] |
| TERC | Dyskeratosis congenita, autosomal dominant 1 | 127550 | Nail dystrophy Lacy skin pigmentation Oral leukoplakia | [185] |
| Gemini of Cajal Body | ||||
| SMN1 | Spinal muscular atrophy, type I Spinal muscular atrophy, type II Spinal muscular atrophy, type III Spinal muscular atrophy, type IV | 253300 253550 253400 271150 | Lower motor neuron degeneration ⬇ SMN protein ⬇ Gems | [177] |
2.2. Nuclear Speckle
2.2.1. Discovery
2.2.2. Key Components
2.2.3. Functions of Key Components
2.2.4. Mechanisms of Gene Expression and the Nuclear Speckle
2.2.5. Human Diseases Associated with the Nuclear Speckle
2.3. Nuclear Stress Body
2.3.1. Discovery
2.3.2. Key Components of the Nuclear Stress Body
2.3.3. Functions of Key Components
| Nuclear Body | Human Homo sapiens | Mouse Mus musculus | African clawed frog Xenopus laevis | Zebrafish Danio rerio | Fruit fly Drosophila melanogaster | Nematode Caenorhabditis elegans | Budding yeast Saccharomyces cerevisiae | References |
|---|---|---|---|---|---|---|---|---|
| Nucleolus | + | + | + | + | + | + | +1 | [211,212,213,214,215] |
| Nuclear speckle | + | + | +2 | ND | + | ND | − | [216,217,218,219] |
| Nuclear stress body | + | − | ND | ND | −3 | −4 | ND | [203,204,205] |
| Transcription factory | + | + | ND | ND | ND | ND | ND | [99,220] |
| Cajal body | + | + | −5 | + | + | ND | +6 | [71,216,221,222,223,224] |
| Gemini of Cajal body | + | + | ND | ND | −7 | ND | ND | [60,225,226] |
| Histone locus body | + | ND | +8 | −9 | + | ND | ND | [71,77,223,227,228] |
| Paraspeckle | + | + | ND | ND | ND | ND | ND | [78,79] |
2.3.4. Mechanisms of Gene Expression and the Nuclear Stress Body
2.3.5. Human Diseases Associated with the Nuclear Stress Body
2.4. Transcription Factory
2.4.1. Discovery
2.4.2. Key Components of the Transcription Factory
2.4.3. Gene Regulatory Functions of Key Components
2.4.4. Mechanisms of Gene Expression and the Transcription Factory
2.4.5. Human Diseases Associated with the Transcription Factory
2.5. Cajal Body
2.5.1. Discovery
2.5.2. Key Components
2.5.3. Functions of Key Components
2.5.4. Mechanisms of Gene Expression and the Cajal Body
2.5.5. Human Diseases Associated with the Cajal Body
2.6. Gemini of Cajal Body
2.6.1. Discovery
2.6.2. Key Components of the Gemini of Cajal Body
2.6.3. Functions of Key Components
2.6.4. Mechanisms of Gene Expression and the Gemini of Cajal Body
2.6.5. Human Diseases Associated with the Gemini of Cajal Body
2.7. Histone Locus Body
2.7.1. Discovery
2.7.2. Key Components of the Histone Locus Body
2.7.3. Functions of Key Components
2.7.4. Mechanisms of Gene Expression and the Histone Locus Body
2.7.5. Human Diseases Associated with the Histone Locus Body
2.8. Paraspeckle
2.8.1. Discovery
2.8.2. Key Components of the Paraspeckle
2.8.3. Functions of Key Components
2.8.4. Mechanisms of Gene Expression and the Paraspeckle
2.8.5. Human Diseases Associated with the Paraspeckle
3. Discussion
4. Conclusion
Acknowledgements
Conflict of Interest
References and Notes
- Venters, B.J.; Pugh, B.F. How eukaryotic genes are transcribed. Crit. Rev. Biochem. Mol. Biol. 2009, 44, 117–141. [Google Scholar]
- Hocine, S.; Singer, R.H.; Grunwald, D. RNA processing and export. Cold Spring Harb. Perspect. Biol. 2010, 2, a000752. [Google Scholar] [CrossRef]
- Gebauer, F.; Hentze, M.W. Molecular mechanisms of translational control. Nat. Rev. Mol. Cell Biol. 2004, 5, 827–835. [Google Scholar] [CrossRef]
- Carmo-Fonseca, M.; Berciano, M.T.; Lafarga, M. Orphan nuclear bodies. Cold Spring Harb. Perspect. Biol. 2010, 2, a000703. [Google Scholar] [CrossRef]
- Lallemand-Breitenbach, V.; de The, H. PML nuclear bodies. Cold Spring Harb. Perspect. Biol. 2010, 2, a000661. [Google Scholar] [CrossRef]
- Wagner, R. Einige bemerkungen und fragen über das keimbläschen (vesicular germinativa). Müller's Arch. Anat. Physiol.Wissenschaftliche Med. 1835, 373–377. [Google Scholar]
- Valentin, G. Repertorium für anatomie und physiologie; Verlag von Veit und Comp: Berlin, Germany, 1836; Volume 1, pp. 1–293. [Google Scholar]
- Brown, D.D.; Gurdon, J.B. Absence of ribosomal RNA synthesis in the anucleolate mutant of Xenopus laevis. Proc. Natl. Acad. Sci. USA 1964, 51, 139–146. [Google Scholar] [CrossRef]
- Boisvert, F.M.; van Koningsbruggen, S.; Navascues, J.; Lamond, A.I. The multifunctional nucleolus. Nat. Rev. Mol. Cell Biol. 2007, 8, 574–585. [Google Scholar] [CrossRef]
- Boulon, S.; Westman, B.J.; Hutten, S.; Boisvert, F.M.; Lamond, A.I. The nucleolus under stress. Mol. Cell 2010, 40, 216–227. [Google Scholar] [CrossRef]
- Gerbi, S.A.; Borovjagin, A.V.; Lange, T.S. The nucleolus: A site of ribonucleoprotein maturation. Curr. Opin. Cell Biol. 2003, 15, 318–325. [Google Scholar] [CrossRef]
- Andersen, J.S.; Lyon, C.E.; Fox, A.H.; Leung, A.K.; Lam, Y.W.; Steen, H.; Mann, M.; Lamond, A.I. Directed proteomic analysis of the human nucleolus. Curr. Biol. 2002, 12, 1–11. [Google Scholar]
- Scherl, A.; Coute, Y.; Deon, C.; Calle, A.; Kindbeiter, K.; Sanchez, J.C.; Greco, A.; Hochstrasser, D.; Diaz, J.J. Functional proteomic analysis of human nucleolus. Mol. Biol. Cell 2002, 13, 4100–4109. [Google Scholar] [CrossRef]
- Maggi, L.B., Jr.; Kuchenruether, M.; Dadey, D.Y.; Schwope, R.M.; Grisendi, S.; Townsend, R.R.; Pandolfi, P.P.; Weber, J.D. Nucleophosmin serves as a rate-limiting nuclear export chaperone for the mammalian ribosome. Mol. Cell. Biol. 2008, 28, 7050–7065. [Google Scholar] [CrossRef]
- Ginisty, H.; Amalric, F.; Bouvet, P. Nucleolin functions in the first step of ribosomal RNA processing. EMBO J. 1998, 17, 1476–1486. [Google Scholar] [CrossRef]
- Rickards, B.; Flint, S.J.; Cole, M.D.; LeRoy, G. Nucleolin is required for RNA polymerase I transcription in vivo. Mol. Cell. Biol. 2007, 27, 937–948. [Google Scholar] [CrossRef]
- Kiss, T. Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs. EMBO J. 2001, 20, 3617–3622. [Google Scholar] [CrossRef]
- Isaac, C.; Yang, Y.; Meier, U.T. Nopp140 functions as a molecular link between the nucleolus and the coiled bodies. J. Cell Biol. 1998, 142, 319–329. [Google Scholar] [CrossRef]
- Spector, D.L.; Ochs, R.L.; Busch, H. Silver staining, immunofluorescence, and immunoelectron microscopic localization of nucleolar phosphoproteins B23 and C2. Chromosoma 1984, 90, 139–148. [Google Scholar] [CrossRef]
- Ochs, R.L.; Lischwe, M.A.; Spohn, W.H.; Busch, H. Fibrillarin: A new protein of the nucleolus identified by autoimmune sera. Biol. Cell 1985, 54, 123–133. [Google Scholar] [CrossRef]
- Leung, A.K.; Lamond, A.I. In vivo analysis of NHPX reveals a novel nucleolar localization pathway involving a transient accumulation in splicing speckles. J. Cell Biol. 2002, 157, 615–629. [Google Scholar] [CrossRef] [Green Version]
- Gautier, T.; Berges, T.; Tollervey, D.; Hurt, E. Nucleolar KKE/D repeat proteins Nop56p and Nop58p interact with Nop1p and are required for ribosome biogenesis. Mol. Cell. Biol. 1997, 17, 7088–7098. [Google Scholar]
- Heiss, N.S.; Girod, A.; Salowsky, R.; Wiemann, S.; Pepperkok, R.; Poustka, A. Dyskerin localizes to the nucleolus and its mislocalization is unlikely to play a role in the pathogenesis of dyskeratosis congenita. Hum. Mol. Genet. 1999, 8, 2515–2524. [Google Scholar] [CrossRef]
- Girard, J.P.; Lehtonen, H.; Caizergues-Ferrer, M.; Amalric, F.; Tollervey, D.; Lapeyre, B. GAR1 is an essential small nucleolar RNP protein required for pre-rRNA processing in yeast. EMBO J. 1992, 11, 673–682. [Google Scholar]
- Henras, A.; Henry, Y.; Bousquet-Antonelli, C.; Noaillac-Depeyre, J.; Gelugne, J.P.; Caizergues-Ferrer, M. Nhp2p and Nop10p are essential for the function of H/ACA snoRNPs. EMBO J. 1998, 17, 7078–7090. [Google Scholar]
- Meier, U.T.; Blobel, G. Nopp140 shuttles on tracks between nucleolus and cytoplasm. Cell 1992, 70, 127–138. [Google Scholar] [CrossRef]
- Scheer, U.; Rose, K.M. Localization of RNA polymerase I in interphase cells and mitotic chromosomes by light and electron microscopic immunocytochemistry. Proc. Natl. Acad. Sci. USA 1984, 81, 1431–1435. [Google Scholar] [CrossRef]
- Tyc, K.; Steitz, J.A. U3, U8 and U13 comprise a new class of mammalian snRNPs localized in the cell nucleolus. EMBO J. 1989, 8, 3113–3119. [Google Scholar]
- Lange, T.S.; Borovjagin, A.; Maxwell, E.S.; Gerbi, S.A. Conserved boxes C and D are essential nucleolar localization elements of U14 and U8 snoRNAs. EMBO J. 1998, 17, 3176–3187. [Google Scholar] [CrossRef]
- Selvamurugan, N.; Joost, O.H.; Haas, E.S.; Brown, J.W.; Galvin, N.J.; Eliceiri, G.L. Intracellular localization and unique conserved sequences of three small nucleolar RNAs. Nucleic Acids Res. 1997, 25, 1591–1596. [Google Scholar] [CrossRef]
- Caceres, J.F.; Misteli, T.; Screaton, G.R.; Spector, D.L.; Krainer, A.R. Role of the modular domains of SR proteins in subnuclear localization and alternative splicing specificity. J. Cell Biol. 1997, 138, 225–238. [Google Scholar] [CrossRef] [Green Version]
- Fu, X.D.; Maniatis, T. Factor required for mammalian spliceosome assembly is localized to discrete regions in the nucleus. Nature 1990, 343, 437–441. [Google Scholar] [CrossRef]
- Mintz, P.J.; Patterson, S.D.; Neuwald, A.F.; Spahr, C.S.; Spector, D.L. Purification and biochemical characterization of interchromatin granule clusters. EMBO J. 1999, 18, 4308–4320. [Google Scholar]
- Saitoh, N.; Spahr, C.S.; Patterson, S.D.; Bubulya, P.; Neuwald, A.F.; Spector, D.L. Proteomic analysis of interchromatin granule clusters. Mol. Biol. Cell 2004, 15, 3876–3890. [Google Scholar] [CrossRef]
- Colwill, K.; Pawson, T.; Andrews, B.; Prasad, J.; Manley, J.L.; Bell, J.C.; Duncan, P.I. The Clk/Sty protein kinase phosphorylates SR splicing factors and regulates their intranuclear distribution. EMBO J. 1996, 15, 265–275. [Google Scholar]
- Bregman, D.B.; Du, L.; van der Zee, S.; Warren, S.L. Transcription-dependent redistribution of the large subunit of RNA polymerase II to discrete nuclear domains. J. Cell Biol. 1995, 129, 287–298. [Google Scholar] [CrossRef]
- Huang, S.; Spector, D.L. U1 and U2 small nuclear RNAs are present in nuclear speckles. Proc. Natl. Acad. Sci. USA 1992, 89, 305–308. [Google Scholar] [CrossRef]
- Hutchinson, J.N.; Ensminger, A.W.; Clemson, C.M.; Lynch, C.R.; Lawrence, J.B.; Chess, A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics 2007, 8, 39. [Google Scholar]
- Carter, K.C.; Taneja, K.L.; Lawrence, J.B. Discrete nuclear domains of poly(A) RNA and their relationship to the functional organization of the nucleus. J. Cell Biol. 1991, 115, 1191–1202. [Google Scholar] [CrossRef]
- Sarge, K.D.; Murphy, S.P.; Morimoto, R.I. Activation of heat shock gene transcription by heat shock factor 1 involves oligomerization, acquisition of DNA-binding activity, and nuclear localization and can occur in the absence of stress. Mol. Cell. Biol. 1993, 13, 1392–1407. [Google Scholar]
- Alastalo, T.P.; Hellesuo, M.; Sandqvist, A.; Hietakangas, V.; Kallio, M.; Sistonen, L. Formation of nuclear stress granules involves HSF2 and coincides with the nucleolar localization of Hsp70. J. Cell Sci. 2003, 116, 3557–3570. [Google Scholar] [CrossRef]
- Weighardt, F.; Cobianchi, F.; Cartegni, L.; Chiodi, I.; Villa, A.; Riva, S.; Biamonti, G. A novel hnRNP protein (HAP/SAF-B) enters a subset of hnRNP complexes and relocates in nuclear granules in response to heat shock. J. Cell Sci. 1999, 112, 1465–1476. [Google Scholar]
- Denegri, M.; Chiodi, I.; Corioni, M.; Cobianchi, F.; Riva, S.; Biamonti, G. Stress-induced nuclear bodies are sites of accumulation of pre-mRNA processing factors. Mol. Biol. Cell 2001, 12, 3502–3514. [Google Scholar] [CrossRef]
- Jolly, C.; Metz, A.; Govin, J.; Vigneron, M.; Turner, B.M.; Khochbin, S.; Vourc'h, C. Stress-induced transcription of satellite III repeats. J. Cell Biol. 2004, 164, 25–33. [Google Scholar] [CrossRef]
- Jackson, D.A.; Iborra, F.J.; Manders, E.M.; Cook, P.R. Numbers and organization of RNA polymerases, nascent transcripts, and transcription units in HeLa nuclei. Mol. Biol. Cell 1998, 9, 1523–1536. [Google Scholar] [CrossRef]
- Melnik, S.; Deng, B.; Papantonis, A.; Baboo, S.; Carr, I.M.; Cook, P.R. The proteomes of transcription factories containing RNA polymerases I, II or III. Nat. Methods 2011, 8, 963–968. [Google Scholar] [CrossRef]
- Raska, I.; Andrade, L.E.; Ochs, R.L.; Chan, E.K.; Chang, C.M.; Roos, G.; Tan, E.M. Immunological and ultrastructural studies of the nuclear coiled body with autoimmune antibodies. Exp. Cell Res. 1991, 195, 27–37. [Google Scholar] [CrossRef]
- Andrade, L.E.; Chan, E.K.; Raska, I.; Peebles, C.L.; Roos, G.; Tan, E.M. Human autoantibody to a novel protein of the nuclear coiled body: Immunological characterization and cDNA cloning of p80-coilin. J. Exp. Med. 1991, 173, 1407–1419. [Google Scholar] [CrossRef]
- Raska, I.; Ochs, R.L.; Andrade, L.E.; Chan, E.K.; Burlingame, R.; Peebles, C.; Gruol, D.; Tan, E.M. Association between the nucleolus and the coiled body. J. Struct. Biol. 1990, 104, 120–127. [Google Scholar] [CrossRef]
- Verheggen, C.; Lafontaine, D.L.; Samarsky, D.; Mouaikel, J.; Blanchard, J.M.; Bordonne, R.; Bertrand, E. Mammalian and yeast U3 snoRNPs are matured in specific and related nuclear compartments. EMBO J. 2002, 21, 2736–2745. [Google Scholar] [CrossRef]
- Meier, U.T.; Blobel, G. NAP57, a mammalian nucleolar protein with a putative homolog in yeast and bacteria. J. Cell Biol. 1994, 127, 1505–1514. [Google Scholar] [CrossRef]
- Pogacic, V.; Dragon, F.; Filipowicz, W. Human H/ACA small nucleolar RNPs and telomerase share evolutionarily conserved proteins NHP2 and NOP10. Mol. Cell. Biol. 2000, 20, 9028–9040. [Google Scholar] [CrossRef]
- Zhu, Y.; Tomlinson, R.L.; Lukowiak, A.A.; Terns, R.M.; Terns, M.P. Telomerase RNA accumulates in Cajal bodies in human cancer cells. Mol. Biol. Cell 2004, 15, 81–90. [Google Scholar]
- Venteicher, A.S.; Abreu, E.B.; Meng, Z.; McCann, K.E.; Terns, R.M.; Veenstra, T.D.; Terns, M.P.; Artandi, S.E. A human telomerase holoenzyme protein required for Cajal body localization and telomere synthesis. Science 2009, 323, 644–648. [Google Scholar] [CrossRef]
- Carvalho, T.; Almeida, F.; Calapez, A.; Lafarga, M.; Berciano, M.T.; Carmo-Fonseca, M. The spinal muscular atrophy disease gene product, SMN: A link between snRNP biogenesis and the Cajal (coiled) body. J. Cell Biol. 1999, 147, 715–728. [Google Scholar] [CrossRef]
- Darzacq, X.; Jady, B.E.; Verheggen, C.; Kiss, A.M.; Bertrand, E.; Kiss, T. Cajal body-specific small nuclear RNAs: A novel class of 2'-O-methylation and pseudouridylation guide RNAs. EMBO J. 2002, 21, 2746–2756. [Google Scholar] [CrossRef]
- Carmo-Fonseca, M.; Pepperkok, R.; Carvalho, M.T.; Lamond, A.I. Transcription-dependent colocalization of the U1, U2, U4/U6, and U5 snRNPs in coiled bodies. J. Cell Biol. 1992, 117, 1–14. [Google Scholar] [CrossRef]
- Matera, A.G.; Ward, D.C. Nucleoplasmic organization of small nuclear ribonucleoproteins in cultured human cells. J. Cell Biol. 1993, 121, 715–727. [Google Scholar] [CrossRef]
- Narayanan, A.; Speckmann, W.; Terns, R.; Terns, M.P. Role of the box C/D motif in localization of small nucleolar RNAs to coiled bodies and nucleoli. Mol. Biol. Cell 1999, 10, 2131–2147. [Google Scholar] [CrossRef]
- Liu, Q.; Dreyfuss, G. A novel nuclear structure containing the survival of motor neurons protein. EMBO J. 1996, 15, 3555–3565. [Google Scholar]
- Liu, Q.; Fischer, U.; Wang, F.; Dreyfuss, G. The spinal muscular atrophy disease gene product, SMN, and its associated protein SIP1 are in a complex with spliceosomal snRNP proteins. Cell 1997, 90, 1013–1021. [Google Scholar] [CrossRef]
- Charroux, B.; Pellizzoni, L.; Perkinson, R.A.; Shevchenko, A.; Mann, M.; Dreyfuss, G. Gemin3: A novel DEAD box protein that interacts with SMN, the spinal muscular atrophy gene product, and is a component of gems. J. Cell Biol. 1999, 147, 1181–1194. [Google Scholar] [CrossRef]
- Charroux, B.; Pellizzoni, L.; Perkinson, R.A.; Yong, J.; Shevchenko, A.; Mann, M.; Dreyfuss, G. Gemin4: A novel component of the SMN complex that is found in both gems and nucleoli. J. Cell Biol. 2000, 148, 1177–1186. [Google Scholar] [CrossRef]
- Gubitz, A.K.; Mourelatos, Z.; Abel, L.; Rappsilber, J.; Mann, M.; Dreyfuss, G. Gemin5, a novel WD repeat protein component of the SMN complex that binds Sm proteins. J. Biol. Chem. 2002, 277, 5631–5636. [Google Scholar]
- Pellizzoni, L.; Baccon, J.; Rappsilber, J.; Mann, M.; Dreyfuss, G. Purification of native survival of motor neurons complexes and identification of Gemin6 as a novel component. J. Biol. Chem. 2002, 277, 7540–7545. [Google Scholar]
- Baccon, J.; Pellizzoni, L.; Rappsilber, J.; Mann, M.; Dreyfuss, G. Identification and characterization of Gemin7, a novel component of the survival of motor neuron complex. J. Biol. Chem. 2002, 277, 31957–31962. [Google Scholar]
- Carissimi, C.; Saieva, L.; Baccon, J.; Chiarella, P.; Maiolica, A.; Sawyer, A.; Rappsilber, J.; Pellizzoni, L. Gemin8 is a novel component of the survival motor neuron complex and functions in small nuclear ribonucleoprotein assembly. J. Biol. Chem. 2006, 281, 8126–8134. [Google Scholar] [CrossRef]
- Gangwani, L.; Mikrut, M.; Theroux, S.; Sharma, M.; Davis, R.J. Spinal muscular atrophy disrupts the interaction of ZPR1 with the SMN protein. Nat. Cell Biol. 2001, 3, 376–383. [Google Scholar] [CrossRef]
- Ma, T.; Van Tine, B.A.; Wei, Y.; Garrett, M.D.; Nelson, D.; Adams, P.D.; Wang, J.; Qin, J.; Chow, L.T.; Harper, J.W. Cell cycle-regulated phosphorylation of p220(NPAT) by cyclin E/Cdk2 in Cajal bodies promotes histone gene transcription. Genes Dev. 2000, 14, 2298–2313. [Google Scholar] [CrossRef]
- Ghule, P.N.; Dominski, Z.; Yang, X.C.; Marzluff, W.F.; Becker, K.A.; Harper, J.W.; Lian, J.B.; Stein, J.L.; van Wijnen, A.J.; Stein, G.S. Staged assembly of histone gene expression machinery at subnuclear foci in the abbreviated cell cycle of human embryonic stem cells. Proc. Natl. Acad. Sci. USA 2008, 105, 16964–16969. [Google Scholar] [CrossRef]
- Liu, J.L.; Murphy, C.; Buszczak, M.; Clatterbuck, S.; Goodman, R.; Gall, J.G. The Drosophila melanogaster Cajal body. J. Cell Biol. 2006, 172, 875–884. [Google Scholar] [CrossRef]
- Barcaroli, D.; Dinsdale, D.; Neale, M.H.; Bongiorno-Borbone, L.; Ranalli, M.; Munarriz, E.; Sayan, A.E.; McWilliam, J.M.; Smith, T.M.; Fava, E.; et al. FLASH is an essential component of Cajal bodies. Proc. Natl. Acad. Sci. USA 2006, 103, 14802–14807. [Google Scholar] [CrossRef]
- Narita, T.; Yung, T.M.; Yamamoto, J.; Tsuboi, Y.; Tanabe, H.; Tanaka, K.; Yamaguchi, Y.; Handa, H. NELF interacts with CBC and participates in 3' end processing of replication-dependent histone mRNAs. Mol. Cell 2007, 26, 349–365. [Google Scholar] [CrossRef]
- Miele, A.; Braastad, C.D.; Holmes, W.F.; Mitra, P.; Medina, R.; Xie, R.; Zaidi, S.K.; Ye, X.; Wei, Y.; Harper, J.W.; et al. HiNF-P directly links the cyclin E/CDK2/p220NPAT pathway to histone H4 gene regulation at the G1/S phase cell cycle transition. Mol. Cell. Biol. 2005, 25, 6140–6153. [Google Scholar] [CrossRef]
- Gangwani, L. Deficiency of the zinc finger protein ZPR1 causes defects in transcription and cell cycle progression. J. Biol. Chem. 2006, 281, 40330–40340. [Google Scholar] [CrossRef]
- Liu, J.L.; Wu, Z.; Nizami, Z.; Deryusheva, S.; Rajendra, T.K.; Beumer, K.J.; Gao, H.; Matera, A.G.; Carroll, D.; Gall, J.G. Coilin is essential for Cajal body organization in Drosophila melanogaster. Mol. Biol. Cell 2009, 20, 1661–1670. [Google Scholar] [CrossRef]
- Frey, M.R.; Matera, A.G. Coiled bodies contain U7 small nuclear RNA and associate with specific DNA sequences in interphase human cells. Proc. Natl. Acad. Sci. USA 1995, 92, 5915–5919. [Google Scholar] [CrossRef]
- Fox, A.H.; Lam, Y.W.; Leung, A.K.; Lyon, C.E.; Andersen, J.; Mann, M.; Lamond, A.I. Paraspeckles: A novel nuclear domain. Curr. Biol. 2002, 12, 13–25. [Google Scholar] [CrossRef]
- Prasanth, K.V.; Prasanth, S.G.; Xuan, Z.; Hearn, S.; Freier, S.M.; Bennett, C.F.; Zhang, M.Q.; Spector, D.L. Regulating gene expression through RNA nuclear retention. Cell 2005, 123, 249–263. [Google Scholar] [CrossRef]
- Dettwiler, S.; Aringhieri, C.; Cardinale, S.; Keller, W.; Barabino, S.M. Distinct sequence motifs within the 68-kDa subunit of cleavage factor Im mediate RNA binding, protein-protein interactions, and subcellular localization. J. Biol. Chem. 2004, 279, 35788–35797. [Google Scholar] [CrossRef]
- Clemson, C.M.; Hutchinson, J.N.; Sara, S.A.; Ensminger, A.W.; Fox, A.H.; Chess, A.; Lawrence, J.B. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol. Cell 2009, 33, 717–726. [Google Scholar] [CrossRef]
- Sunwoo, H.; Dinger, M.E.; Wilusz, J.E.; Amaral, P.P.; Mattick, J.S.; Spector, D.L. MEN epsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. 2009, 19, 347–359. [Google Scholar]
- Sasaki, Y.T.; Ideue, T.; Sano, M.; Mituyama, T.; Hirose, T. MENepsilon/beta noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc. Natl. Acad. Sci. USA 2009, 106, 2525–2530. [Google Scholar]
- Tollervey, D.; Lehtonen, H.; Carmo-Fonseca, M.; Hurt, E.C. The small nucleolar RNP protein NOP1 (fibrillarin) is required for pre-rRNA processing in yeast. EMBO J. 1991, 10, 573–583. [Google Scholar]
- Jack, K.; Bellodi, C.; Landry, D.M.; Niederer, R.O.; Meskauskas, A.; Musalgaonkar, S.; Kopmar, N.; Krasnykh, O.; Dean, A.M.; Thompson, S.R.; et al. rRNA pseudouridylation defects affect ribosomal ligand binding and translational fidelity from yeast to human cells. Mol. Cell 2011, 44, 660–666. [Google Scholar] [CrossRef]
- Haaf, T.; Ward, D.C. Inhibition of RNA polymerase II transcription causes chromatin decondensation, loss of nucleolar structure, and dispersion of chromosomal domains. Exp. Cell Res. 1996, 224, 163–173. [Google Scholar] [CrossRef]
- Flygare, J.; Aspesi, A.; Bailey, J.C.; Miyake, K.; Caffrey, J.M.; Karlsson, S.; Ellis, S.R. Human RPS19, the gene mutated in Diamond-Blackfan anemia, encodes a ribosomal protein required for the maturation of 40S ribosomal subunits. Blood 2007, 109, 980–986. [Google Scholar]
- Hughes, J.M.; Ares, M., Jr. Depletion of U3 small nucleolar RNA inhibits cleavage in the 5' external transcribed spacer of yeast pre-ribosomal RNA and impairs formation of 18S ribosomal RNA. EMBO J. 1991, 10, 4231–4239. [Google Scholar]
- Tripathi, V.; Song, D.Y.; Zong, X.; Shevtsov, S.P.; Hearn, S.; Fu, X.D.; Dundr, M.; Prasanth, K.V. SRSF1 regulates the assembly of pre-mRNA processing factors in nuclear speckles. Mol. Biol. Cell 2012, 23, 3694–3706. [Google Scholar]
- Sacco-Bubulya, P.; Spector, D.L. Disassembly of interchromatin granule clusters alters the coordination of transcription and pre-mRNA splicing. J. Cell Biol. 2002, 156, 425–436. [Google Scholar] [CrossRef]
- Tripathi, V.; Ellis, J.D.; Shen, Z.; Song, D.Y.; Pan, Q.; Watt, A.T.; Freier, S.M.; Bennett, C.F.; Sharma, A.; Bubulya, P.A.; et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell 2010, 39, 925–938. [Google Scholar] [CrossRef]
- Sharma, A.; Takata, H.; Shibahara, K.; Bubulya, A.; Bubulya, P.A. Son is essential for nuclear speckle organization and cell cycle progression. Mol. Biol. Cell 2010, 21, 650–663. [Google Scholar] [CrossRef]
- Okada, M.; Jang, S.W.; Ye, K. Akt phosphorylation and nuclear phosphoinositide association mediate mRNA export and cell proliferation activities by ALY. Proc. Natl. Acad. Sci. USA 2008, 105, 8649–8654. [Google Scholar] [CrossRef]
- Dias, A.P.; Dufu, K.; Lei, H.; Reed, R. A role for TREX components in the release of spliced mRNA from nuclear speckle domains. Nat. Commun. 2010, 1, 97. [Google Scholar] [CrossRef]
- Nakagawa, S.; Ip, J.Y.; Shioi, G.; Tripathi, V.; Zong, X.; Hirose, T.; Prasanth, K.V. Malat1 is not an essential component of nuclear speckles in mice. RNA 2012, 18, 1487–1499. [Google Scholar] [CrossRef]
- Eissmann, M.; Gutschner, T.; Hammerle, M.; Gunther, S.; Caudron-Herger, M.; Gross, M.; Schirmacher, P.; Rippe, K.; Braun, T.; Zornig, M.; et al. Loss of the abundant nuclear non-coding RNA MALAT1 is compatible with life and development. RNA Biol. 2012, 9, 1076–1087. [Google Scholar]
- O'Keefe, R.T.; Mayeda, A.; Sadowski, C.L.; Krainer, A.R.; Spector, D.L. Disruption of pre-mRNA splicing in vivo results in reorganization of splicing factors. J. Cell Biol. 1994, 124, 249–260. [Google Scholar] [CrossRef]
- Sandqvist, A.; Bjork, J.K.; Akerfelt, M.; Chitikova, Z.; Grichine, A.; Vourc'h, C.; Jolly, C.; Salminen, T.A.; Nymalm, Y.; Sistonen, L. Heterotrimerization of heat-shock factors 1 and 2 provides a transcriptional switch in response to distinct stimuli. Mol. Biol. Cell 2009, 20, 1340–1347. [Google Scholar] [CrossRef]
- Jackson, D.A.; Hassan, A.B.; Errington, R.J.; Cook, P.R. Visualization of focal sites of transcription within human nuclei. EMBO J. 1993, 12, 1059–1065. [Google Scholar]
- Pombo, A.; Jackson, D.A.; Hollinshead, M.; Wang, Z.; Roeder, R.G.; Cook, P.R. Regional specialization in human nuclei: Visualization of discrete sites of transcription by RNA polymerase III. EMBO J. 1999, 18, 2241–2253. [Google Scholar] [CrossRef]
- Tucker, K.E.; Berciano, M.T.; Jacobs, E.Y.; LePage, D.F.; Shpargel, K.B.; Rossire, J.J.; Chan, E.K.; Lafarga, M.; Conlon, R.A.; Matera, A.G. Residual Cajal bodies in coilin knockout mice fail to recruit Sm snRNPs and SMN, the spinal muscular atrophy gene product. J. Cell Biol. 2001, 154, 293–307. [Google Scholar] [CrossRef]
- Walker, M.P.; Tian, L.; Matera, A.G. Reduced viability, fertility and fecundity in mice lacking the cajal body marker protein, coilin. PLoS One 2009, 4, e6171. [Google Scholar] [CrossRef]
- Bauer, D.W.; Gall, J.G. Coiled bodies without coilin. Mol. Biol. Cell 1997, 8, 73–82. [Google Scholar]
- Deryusheva, S.; Gall, J.G. Small Cajal body-specific RNAs of Drosophila function in the absence of Cajal bodies. Mol. Biol. Cell 2009, 20, 5250–5259. [Google Scholar] [CrossRef]
- Batista, L.F.; Pech, M.F.; Zhong, F.L.; Nguyen, H.N.; Xie, K.T.; Zaug, A.J.; Crary, S.M.; Choi, J.; Sebastiano, V.; Cherry, A.; et al. Telomere shortening and loss of self-renewal in dyskeratosis congenita induced pluripotent stem cells. Nature 2011, 474, 399–402. [Google Scholar] [CrossRef]
- Renvoise, B.; Colasse, S.; Burlet, P.; Viollet, L.; Meier, U.T.; Lefebvre, S. The loss of the snoRNP chaperone Nopp140 from Cajal bodies of patient fibroblasts correlates with the severity of spinal muscular atrophy. Hum. Mol. Genet. 2009, 18, 1181–1189. [Google Scholar] [CrossRef]
- Lefebvre, S.; Burlet, P.; Liu, Q.; Bertrandy, S.; Clermont, O.; Munnich, A.; Dreyfuss, G.; Melki, J. Correlation between severity and SMN protein level in spinal muscular atrophy. Nat. Genet. 1997, 16, 265–269. [Google Scholar]
- Coovert, D.D.; Le, T.T.; McAndrew, P.E.; Strasswimmer, J.; Crawford, T.O.; Mendell, J.R.; Coulson, S.E.; Androphy, E.J.; Prior, T.W.; Burghes, A.H. The survival motor neuron protein in spinal muscular atrophy. Hum. Mol. Genet. 1997, 6, 1205–1214. [Google Scholar]
- Girard, C.; Neel, H.; Bertrand, E.; Bordonne, R. Depletion of SMN by RNA interference in HeLa cells induces defects in Cajal body formation. Nucleic Acids Res. 2006, 34, 2925–2932. [Google Scholar] [CrossRef]
- Feng, W.; Gubitz, A.K.; Wan, L.; Battle, D.J.; Dostie, J.; Golembe, T.J.; Dreyfuss, G. Gemins modulate the expression and activity of the SMN complex. Hum. Mol. Genet. 2005, 14, 1605–1611. [Google Scholar]
- Shpargel, K.B.; Matera, A.G. Gemin proteins are required for efficient assembly of Sm-class ribonucleoproteins. Proc. Natl. Acad. Sci. USA 2005, 102, 17372–17377. [Google Scholar] [CrossRef]
- Frugier, T.; Tiziano, F.D.; Cifuentes-Diaz, C.; Miniou, P.; Roblot, N.; Dierich, A.; Le Meur, M.; Melki, J. Nuclear targeting defect of SMN lacking the C-terminus in a mouse model of spinal muscular atrophy. Hum. Mol. Genet. 2000, 9, 849–858. [Google Scholar] [CrossRef]
- Gabanella, F.; Butchbach, M.E.; Saieva, L.; Carissimi, C.; Burghes, A.H.; Pellizzoni, L. Ribonucleoprotein assembly defects correlate with spinal muscular atrophy severity and preferentially affect a subset of spliceosomal snRNPs. PLoS One 2007, 2, e921. [Google Scholar]
- Zhang, Z.; Lotti, F.; Dittmar, K.; Younis, I.; Wan, L.; Kasim, M.; Dreyfuss, G. SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing. Cell 2008, 133, 585–600. [Google Scholar]
- Campion, Y.; Neel, H.; Gostan, T.; Soret, J.; Bordonne, R. Specific splicing defects in S. pombe carrying a degron allele of the Survival of Motor Neuron gene. EMBO J. 2010, 29, 1817–1829. [Google Scholar] [CrossRef]
- Shpargel, K.B.; Praveen, K.; Rajendra, T.K.; Matera, A.G. Gemin3 is an essential gene required for larval motor function and pupation in Drosophila. Mol. Biol. Cell 2009, 20, 90–101. [Google Scholar]
- Gangwani, L.; Flavell, R.A.; Davis, R.J. ZPR1 is essential for survival and is required for localization of the survival motor neurons (SMN) protein to Cajal bodies. Mol. Cell. Biol. 2005, 25, 2744–2756. [Google Scholar] [CrossRef]
- Doran, B.; Gherbesi, N.; Hendricks, G.; Flavell, R.A.; Davis, R.J.; Gangwani, L. Deficiency of the zinc finger protein ZPR1 causes neurodegeneration. Proc. Natl. Acad. Sci. USA 2006, 103, 7471–7475. [Google Scholar]
- Di Fruscio, M.; Weiher, H.; Vanderhyden, B.C.; Imai, T.; Shiomi, T.; Hori, T.A.; Jaenisch, R.; Gray, D.A. Proviral inactivation of the Npat gene of Mpv 20 mice results in early embryonic arrest. Mol. Cell. Biol. 1997, 17, 4080–4086. [Google Scholar]
- Sullivan, K.D.; Mullen, T.E.; Marzluff, W.F.; Wagner, E.J. Knockdown of SLBP results in nuclear retention of histone mRNA. RNA 2009, 15, 459–472. [Google Scholar] [CrossRef]
- Sullivan, E.; Santiago, C.; Parker, E.D.; Dominski, Z.; Yang, X.; Lanzotti, D.J.; Ingledue, T.C.; Marzluff, W.F.; Duronio, R.J. Drosophila stem loop binding protein coordinates accumulation of mature histone mRNA with cell cycle progression. Genes Dev. 2001, 15, 173–187. [Google Scholar] [CrossRef]
- White, A.E.; Leslie, M.E.; Calvi, B.R.; Marzluff, W.F.; Duronio, R.J. Developmental and cell cycle regulation of the Drosophila histone locus body. Mol. Biol. Cell 2007, 18, 2491–2502. [Google Scholar] [CrossRef]
- Godfrey, A.C.; White, A.E.; Tatomer, D.C.; Marzluff, W.F.; Duronio, R.J. The Drosophila U7 snRNP proteins Lsm10 and Lsm11 are required for histone pre-mRNA processing and play an essential role in development. RNA 2009, 15, 1661–1672. [Google Scholar] [CrossRef]
- Barcaroli, D.; Bongiorno-Borbone, L.; Terrinoni, A.; Hofmann, T.G.; Rossi, M.; Knight, R.A.; Matera, A.G.; Melino, G.; De Laurenzi, V. FLASH is required for histone transcription and S-phase progression. Proc. Natl. Acad. Sci. USA 2006, 103, 14808–14812. [Google Scholar] [CrossRef]
- De Cola, A.; Bongiorno-Borbone, L.; Bianchi, E.; Barcaroli, D.; Carletti, E.; Knight, R.A.; Di Ilio, C.; Melino, G.; Sette, C.; De Laurenzi, V. FLASH is essential during early embryogenesis and cooperates with p73 to regulate histone gene transcription. Oncogene 2012, 31, 573–582. [Google Scholar]
- Xie, R.; Medina, R.; Zhang, Y.; Hussain, S.; Colby, J.; Ghule, P.; Sundararajan, S.; Keeler, M.; Liu, L.J.; van der Deen, M.; et al. The histone gene activator HINFP is a nonredundant cyclin E/CDK2 effector during early embryonic cell cycles. Proc. Natl. Acad. Sci. USA 2009, 106, 12359–12364. [Google Scholar] [CrossRef]
- Godfrey, A.C.; Kupsco, J.M.; Burch, B.D.; Zimmerman, R.M.; Dominski, Z.; Marzluff, W.F.; Duronio, R.J. U7 snRNA mutations in Drosophila block histone pre-mRNA processing and disrupt oogenesis. RNA 2006, 12, 396–409. [Google Scholar] [CrossRef]
- Salzler, H.R.; Tatomer, D.C.; Malek, P.Y.; McDaniel, S.L.; Orlando, A.N.; Marzluff, W.F.; Duronio, R.J. A Sequence in the Drosophila H3-H4 Promoter Triggers Histone Locus Body Assembly and Biosynthesis of Replication-Coupled Histone mRNAs. Dev. Cell 2013, 24, 623–634. [Google Scholar] [CrossRef]
- Hata, K.; Nishimura, R.; Muramatsu, S.; Matsuda, A.; Matsubara, T.; Amano, K.; Ikeda, F.; Harley, V.R.; Yoneda, T. Paraspeckle protein p54nrb links Sox9-mediated transcription with RNA processing during chondrogenesis in mice. J. Clin. Invest. 2008, 118, 3098–3108. [Google Scholar] [CrossRef]
- Brown, S.A.; Ripperger, J.; Kadener, S.; Fleury-Olela, F.; Vilbois, F.; Rosbash, M.; Schibler, U. PERIOD1-associated proteins modulate the negative limb of the mammalian circadian oscillator. Science 2005, 308, 693–696. [Google Scholar] [CrossRef]
- Chen, L.L.; Carmichael, G.G. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: Functional role of a nuclear noncoding RNA. Mol. Cell 2009, 35, 467–478. [Google Scholar] [CrossRef]
- Kowalska, E.; Ripperger, J.A.; Muheim, C.; Maier, B.; Kurihara, Y.; Fox, A.H.; Kramer, A.; Brown, S.A. Distinct roles of DBHS family members in the circadian transcriptional feedback loop. Mol. Cell. Biol. 2012, 32, 4585–4594. [Google Scholar] [CrossRef]
- Nakagawa, S.; Naganuma, T.; Shioi, G.; Hirose, T. Paraspeckles are subpopulation-specific nuclear bodies that are not essential in mice. J. Cell Biol. 2011, 193, 31–39. [Google Scholar] [CrossRef]
- Murano, K.; Okuwaki, M.; Hisaoka, M.; Nagata, K. Transcription regulation of the rRNA gene by a multifunctional nucleolar protein, B23/nucleophosmin, through its histone chaperone activity. Mol. Cell. Biol. 2008, 28, 3114–3126. [Google Scholar]
- Cong, R.; Das, S.; Ugrinova, I.; Kumar, S.; Mongelard, F.; Wong, J.; Bouvet, P. Interaction of nucleolin with ribosomal RNA genes and its role in RNA polymerase I transcription. Nucleic Acids Res. 2012, 40, 9441–9454. [Google Scholar]
- Lessard, F.; Stefanovsky, V.; Tremblay, M.G.; Moss, T. The cellular abundance of the essential transcription termination factor TTF-I regulates ribosome biogenesis and is determined by MDM2 ubiquitinylation. Nucleic Acids Res. 2012, 40, 5357–5367. [Google Scholar] [CrossRef]
- Boyd, M.T.; Vlatkovic, N.; Rubbi, C.P. The nucleolus directly regulates p53 export and degradation. J. Cell Biol. 2011, 194, 689–703. [Google Scholar]
- Richardson, L.A.; Reed, B.J.; Charette, J.M.; Freed, E.F.; Fredrickson, E.K.; Locke, M.N.; Baserga, S.J.; Gardner, R.G. A conserved deubiquitinating enzyme controls cell growth by regulating RNA polymerase I stability. Cell Rep. 2012, 2, 372–385. [Google Scholar] [CrossRef]
- Westman, B.J.; Verheggen, C.; Hutten, S.; Lam, Y.W.; Bertrand, E.; Lamond, A.I. A proteomic screen for nucleolar SUMO targets shows SUMOylation modulates the function of Nop5/Nop58. Mol. Cell 2010, 39, 618–631. [Google Scholar] [CrossRef]
- Haindl, M.; Harasim, T.; Eick, D.; Muller, S. The nucleolar SUMO-specific protease SENP3 reverses SUMO modification of nucleophosmin and is required for rRNA processing. EMBO Rep. 2008, 9, 273–279. [Google Scholar] [CrossRef]
- Bernardi, R.; Scaglioni, P.P.; Bergmann, S.; Horn, H.F.; Vousden, K.H.; Pandolfi, P.P. PML regulates p53 stability by sequestering Mdm2 to the nucleolus. Nat. Cell Biol. 2004, 6, 665–672. [Google Scholar] [CrossRef]
- Mekhail, K.; Gunaratnam, L.; Bonicalzi, M.E.; Lee, S. HIF activation by pH-dependent nucleolar sequestration of VHL. Nat. Cell Biol. 2004, 6, 642–647. [Google Scholar]
- Fatyol, K.; Szalay, A.A. The p14ARF tumor suppressor protein facilitates nucleolar sequestration of hypoxia-inducible factor-1alpha (HIF-1alpha ) and inhibits HIF-1-mediated transcription. J. Biol. Chem. 2001, 276, 28421–28429. [Google Scholar] [CrossRef]
- Lin, D.Y.; Shih, H.M. Essential role of the 58-kDa microspherule protein in the modulation of Daxx-dependent transcriptional repression as revealed by nucleolar sequestration. J. Biol. Chem. 2002, 277, 25446–25456. [Google Scholar]
- Bywater, M.J.; Pearson, R.B.; McArthur, G.A.; Hannan, R.D. Dysregulation of the basal RNA polymerase transcription apparatus in cancer. Nat. Rev. Cancer 2013, 13, 299–314. [Google Scholar]
- The Treacher Collins Syndrome Collaborative Group. Positional cloning of a gene involved in the pathogenesis of Treacher Collins syndrome. Nat. Genet 1996, 12, 130–136. [CrossRef]
- Dauwerse, J.G.; Dixon, J.; Seland, S.; Ruivenkamp, C.A.; van Haeringen, A.; Hoefsloot, L.H.; Peters, D.J.; Boers, A.C.; Daumer-Haas, C.; Maiwald, R.; et al. Mutations in genes encoding subunits of RNA polymerases I and III cause Treacher Collins syndrome. Nat. Genet. 2011, 43, 20–22. [Google Scholar]
- Draptchinskaia, N.; Gustavsson, P.; Andersson, B.; Pettersson, M.; Willig, T.N.; Dianzani, I.; Ball, S.; Tchernia, G.; Klar, J.; Matsson, H.; et al. The gene encoding ribosomal protein S19 is mutated in Diamond-Blackfan anaemia. Nat. Genet. 1999, 21, 169–175. [Google Scholar]
- Gazda, H.T.; Grabowska, A.; Merida-Long, L.B.; Latawiec, E.; Schneider, H.E.; Lipton, J.M.; Vlachos, A.; Atsidaftos, E.; Ball, S.E.; Orfali, K.A.; et al. Ribosomal protein S24 gene is mutated in Diamond-Blackfan anemia. Am. J. Hum. Genet. 2006, 79, 1110–1118. [Google Scholar] [CrossRef]
- Cmejla, R.; Cmejlova, J.; Handrkova, H.; Petrak, J.; Pospisilova, D. Ribosomal protein S17 gene (RPS17) is mutated in Diamond-Blackfan anemia. Hum. Mutat. 2007, 28, 1178–1182. [Google Scholar] [CrossRef]
- Farrar, J.E.; Nater, M.; Caywood, E.; McDevitt, M.A.; Kowalski, J.; Takemoto, C.M.; Talbot, C.C., Jr.; Meltzer, P.; Esposito, D.; Beggs, A.H.; et al. Abnormalities of the large ribosomal subunit protein, Rpl35a, in Diamond-Blackfan anemia. Blood 2008, 112, 1582–1592. [Google Scholar] [CrossRef]
- Gazda, H.T.; Sheen, M.R.; Vlachos, A.; Choesmel, V.; O'Donohue, M.F.; Schneider, H.; Darras, N.; Hasman, C.; Sieff, C.A.; Newburger, P.E.; et al. Ribosomal protein L5 and L11 mutations are associated with cleft palate and abnormal thumbs in Diamond-Blackfan anemia patients. Am. J. Hum. Genet. 2008, 83, 769–780. [Google Scholar] [CrossRef]
- Doherty, L.; Sheen, M.R.; Vlachos, A.; Choesmel, V.; O'Donohue, M.F.; Clinton, C.; Schneider, H.E.; Sieff, C.A.; Newburger, P.E.; Ball, S.E.; et al. Ribosomal protein genes RPS10 and RPS26 are commonly mutated in Diamond-Blackfan anemia. Am. J. Hum. Genet. 2010, 86, 222–228. [Google Scholar] [CrossRef]
- Gazda, H.T.; Preti, M.; Sheen, M.R.; O'Donohue, M.F.; Vlachos, A.; Davies, S.M.; Kattamis, A.; Doherty, L.; Landowski, M.; Buros, C.; et al. Frameshift mutation in p53 regulator RPL26 is associated with multiple physical abnormalities and a specific pre-ribosomal RNA processing defect in diamond-blackfan anemia. Hum. Mutat. 2012, 33, 1037–1044. [Google Scholar] [CrossRef]
- Yu, C.E.; Oshima, J.; Fu, Y.H.; Wijsman, E.M.; Hisama, F.; Alisch, R.; Matthews, S.; Nakura, J.; Miki, T.; Ouais, S.; et al. Positional cloning of the Werner's syndrome gene. Science 1996, 272, 258–262. [Google Scholar]
- Ellis, N.A.; Groden, J.; Ye, T.Z.; Straughen, J.; Lennon, D.J.; Ciocci, S.; Proytcheva, M.; German, J. The Bloom's syndrome gene product is homologous to RecQ helicases. Cell 1995, 83, 655–666. [Google Scholar] [CrossRef]
- Chakarova, C.F.; Hims, M.M.; Bolz, H.; Abu-Safieh, L.; Patel, R.J.; Papaioannou, M.G.; Inglehearn, C.F.; Keen, T.J.; Willis, C.; Moore, A.T.; et al. Mutations in HPRP3, a third member of pre-mRNA splicing factor genes, implicated in autosomal dominant retinitis pigmentosa. Hum. Mol. Genet. 2002, 11, 87–92. [Google Scholar] [CrossRef]
- Tanackovic, G.; Ransijn, A.; Ayuso, C.; Harper, S.; Berson, E.L.; Rivolta, C. A missense mutation in PRPF6 causes impairment of pre-mRNA splicing and autosomal-dominant retinitis pigmentosa. Am. J. Hum. Genet. 2011, 88, 643–649. [Google Scholar] [CrossRef]
- McKie, A.B.; McHale, J.C.; Keen, T.J.; Tarttelin, E.E.; Goliath, R.; van Lith-Verhoeven, J.J.; Greenberg, J.; Ramesar, R.S.; Hoyng, C.B.; Cremers, F.P.; et al. Mutations in the pre-mRNA splicing factor gene PRPC8 in autosomal dominant retinitis pigmentosa (RP13). Hum. Mol. Genet. 2001, 10, 1555–1562. [Google Scholar] [CrossRef]
- Zhao, C.; Bellur, D.L.; Lu, S.; Zhao, F.; Grassi, M.A.; Bowne, S.J.; Sullivan, L.S.; Daiger, S.P.; Chen, L.J.; Pang, C.P.; et al. Autosomal-dominant retinitis pigmentosa caused by a mutation in SNRNP200, a gene required for unwinding of U4/U6 snRNAs. Am. J. Hum. Genet. 2009, 85, 617–627. [Google Scholar] [CrossRef]
- Lines, M.A.; Huang, L.; Schwartzentruber, J.; Douglas, S.L.; Lynch, D.C.; Beaulieu, C.; Guion-Almeida, M.L.; Zechi-Ceide, R.M.; Gener, B.; Gillessen-Kaesbach, G.; et al. Haploinsufficiency of a spliceosomal GTPase encoded by EFTUD2 causes mandibulofacial dysostosis with microcephaly. Am. J. Hum. Genet. 2012, 90, 369–377. [Google Scholar] [CrossRef]
- Albers, C.A.; Paul, D.S.; Schulze, H.; Freson, K.; Stephens, J.C.; Smethurst, P.A.; Jolley, J.D.; Cvejic, A.; Kostadima, M.; Bertone, P.; et al. Compound inheritance of a low-frequency regulatory SNP and a rare null mutation in exon-junction complex subunit RBM8A causes TAR syndrome. Nat. Genet. 2012, 44, 435–439, S431–S432. [Google Scholar]
- Bernard, G.; Chouery, E.; Putorti, M.L.; Tetreault, M.; Takanohashi, A.; Carosso, G.; Clement, I.; Boespflug-Tanguy, O.; Rodriguez, D.; Delague, V.; et al. Mutations of POLR3A encoding a catalytic subunit of RNA polymerase Pol III cause a recessive hypomyelinating leukodystrophy. Am. J. Hum. Genet. 2011, 89, 415–423. [Google Scholar] [CrossRef]
- Saitsu, H.; Osaka, H.; Sasaki, M.; Takanashi, J.; Hamada, K.; Yamashita, A.; Shibayama, H.; Shiina, M.; Kondo, Y.; Nishiyama, K.; et al. Mutations in POLR3A and POLR3B encoding RNA Polymerase III subunits cause an autosomal-recessive hypomyelinating leukoencephalopathy. Am. J. Hum. Genet. 2011, 89, 644–651. [Google Scholar]
- Tetreault, M.; Choquet, K.; Orcesi, S.; Tonduti, D.; Balottin, U.; Teichmann, M.; Fribourg, S.; Schiffmann, R.; Brais, B.; Vanderver, A.; et al. Recessive mutations in POLR3B, encoding the second largest subunit of Pol III, cause a rare hypomyelinating leukodystrophy. Am. J. Hum. Genet. 2011, 89, 652–655. [Google Scholar] [CrossRef]
- Schwartz, C.E.; Tarpey, P.S.; Lubs, H.A.; Verloes, A.; May, M.M.; Risheg, H.; Friez, M.J.; Futreal, P.A.; Edkins, S.; Teague, J.; et al. The original Lujan syndrome family has a novel missense mutation (p.N1007S) in the MED12 gene. J. Med. Genet. 2007, 44, 472–477. [Google Scholar]
- Risheg, H.; Graham, J.M., Jr.; Clark, R.D.; Rogers, R.C.; Opitz, J.M.; Moeschler, J.B.; Peiffer, A.P.; May, M.; Joseph, S.M.; Jones, J.R.; et al. A recurrent mutation in MED12 leading to R961W causes Opitz-Kaveggia syndrome. Nat. Genet. 2007, 39, 451–453. [Google Scholar]
- Kaufmann, R.; Straussberg, R.; Mandel, H.; Fattal-Valevski, A.; Ben-Zeev, B.; Naamati, A.; Shaag, A.; Zenvirt, S.; Konen, O.; Mimouni-Bloch, A.; et al. Infantile cerebral and cerebellar atrophy is associated with a mutation in the MED17 subunit of the transcription preinitiation mediator complex. Am. J. Hum. Genet. 2010, 87, 667–670. [Google Scholar]
- Hashimoto, S.; Boissel, S.; Zarhrate, M.; Rio, M.; Munnich, A.; Egly, J.M.; Colleaux, L. MED23 mutation links intellectual disability to dysregulation of immediate early gene expression. Science 2011, 333, 1161–1163. [Google Scholar]
- Leal, A.; Huehne, K.; Bauer, F.; Sticht, H.; Berger, P.; Suter, U.; Morera, B.; Del Valle, G.; Lupski, J.R.; Ekici, A.; et al. Identification of the variant Ala335Val of MED25 as responsible for CMT2B2: molecular data, functional studies of the SH3 recognition motif and correlation between wild-type MED25 and PMP22 RNA levels in CMT1A animal models. Neurogenetics 2009. [Google Scholar]
- Arrand, J.E.; Bone, N.M.; Johnson, R.T. Molecular cloning and characterization of a mammalian excision repair gene that partially restores UV resistance to xeroderma pigmentosum complementation group D cells. Proc. Natl. Acad. Sci. USA 1989, 86, 6997–7001. [Google Scholar] [CrossRef]
- Graham, J.M., Jr.; Anyane-Yeboa, K.; Raams, A.; Appeldoorn, E.; Kleijer, W.J.; Garritsen, V.H.; Busch, D.; Edersheim, T.G.; Jaspers, N.G. Cerebro-oculo-facio-skeletal syndrome with a nucleotide excision-repair defect and a mutated XPD gene, with prenatal diagnosis in a triplet pregnancy. Am. J. Hum. Genet. 2001, 69, 291–300. [Google Scholar] [CrossRef]
- Broughton, B.C.; Steingrimsdottir, H.; Weber, C.A.; Lehmann, A.R. Mutations in the xeroderma pigmentosum group D DNA repair/transcription gene in patients with trichothiodystrophy. Nat. Genet. 1994, 7, 189–194. [Google Scholar]
- Weeda, G.; van Ham, R.C.; Vermeulen, W.; Bootsma, D.; van der Eb, A.J.; Hoeijmakers, J.H. A presumed DNA helicase encoded by ERCC-3 is involved in the human repair disorders xeroderma pigmentosum and Cockayne's syndrome. Cell 1990, 62, 777–791. [Google Scholar] [CrossRef]
- Weeda, G.; Eveno, E.; Donker, I.; Vermeulen, W.; Chevallier-Lagente, O.; Taieb, A.; Stary, A.; Hoeijmakers, J.H.; Mezzina, M.; Sarasin, A. A mutation in the XPB/ERCC3 DNA repair transcription gene, associated with trichothiodystrophy. Am. J. Hum. Genet. 1997, 60, 320–329. [Google Scholar]
- Giglia-Mari, G.; Coin, F.; Ranish, J.A.; Hoogstraten, D.; Theil, A.; Wijgers, N.; Jaspers, N.G.; Raams, A.; Argentini, M.; van der Spek, P.J.; et al. A new, tenth subunit of TFIIH is responsible for the DNA repair syndrome trichothiodystrophy group A. Nat. Genet. 2004, 36, 714–719. [Google Scholar]
- Lefebvre, S.; Burglen, L.; Reboullet, S.; Clermont, O.; Burlet, P.; Viollet, L.; Benichou, B.; Cruaud, C.; Millasseau, P.; Zeviani, M.; et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell 1995, 80, 155–165. [Google Scholar] [CrossRef]
- Heiss, N.S.; Knight, S.W.; Vulliamy, T.J.; Klauck, S.M.; Wiemann, S.; Mason, P.J.; Poustka, A.; Dokal, I. X-linked dyskeratosis congenita is caused by mutations in a highly conserved gene with putative nucleolar functions. Nat. Genet. 1998, 19, 32–38. [Google Scholar]
- Knight, S.W.; Heiss, N.S.; Vulliamy, T.J.; Aalfs, C.M.; McMahon, C.; Richmond, P.; Jones, A.; Hennekam, R.C.; Poustka, A.; Mason, P.J.; et al. Unexplained aplastic anaemia, immunodeficiency, and cerebellar hypoplasia (Hoyeraal-Hreidarsson syndrome) due to mutations in the dyskeratosis congenita gene, DKC1. Br. J. Haematol. 1999, 107, 335–339. [Google Scholar] [CrossRef]
- Vulliamy, T.; Beswick, R.; Kirwan, M.; Marrone, A.; Digweed, M.; Walne, A.; Dokal, I. Mutations in the telomerase component NHP2 cause the premature ageing syndrome dyskeratosis congenita. Proc. Natl. Acad. Sci. USA 2008, 105, 8073–8078. [Google Scholar]
- Walne, A.J.; Vulliamy, T.; Marrone, A.; Beswick, R.; Kirwan, M.; Masunari, Y.; Al-Qurashi, F.H.; Aljurf, M.; Dokal, I. Genetic heterogeneity in autosomal recessive dyskeratosis congenita with one subtype due to mutations in the telomerase-associated protein NOP10. Hum. Mol. Genet. 2007, 16, 1619–1629. [Google Scholar] [CrossRef]
- Armanios, M.; Chen, J.L.; Chang, Y.P.; Brodsky, R.A.; Hawkins, A.; Griffin, C.A.; Eshleman, J.R.; Cohen, A.R.; Chakravarti, A.; Hamosh, A.; et al. Haploinsufficiency of telomerase reverse transcriptase leads to anticipation in autosomal dominant dyskeratosis congenita. Proc. Natl. Acad. Sci. USA 2005, 102, 15960–15964. [Google Scholar] [CrossRef]
- Marrone, A.; Walne, A.; Tamary, H.; Masunari, Y.; Kirwan, M.; Beswick, R.; Vulliamy, T.; Dokal, I. Telomerase reverse-transcriptase homozygous mutations in autosomal recessive dyskeratosis congenita and Hoyeraal-Hreidarsson syndrome. Blood 2007, 110, 4198–4205. [Google Scholar] [CrossRef]
- Zhong, F.; Savage, S.A.; Shkreli, M.; Giri, N.; Jessop, L.; Myers, T.; Chen, R.; Alter, B.P.; Artandi, S.E. Disruption of telomerase trafficking by TCAB1 mutation causes dyskeratosis congenita. Genes Dev. 2011, 25, 11–16. [Google Scholar] [CrossRef]
- Vulliamy, T.; Marrone, A.; Goldman, F.; Dearlove, A.; Bessler, M.; Mason, P.J.; Dokal, I. The RNA component of telomerase is mutated in autosomal dominant dyskeratosis congenita. Nature 2001, 413, 432–435. [Google Scholar] [CrossRef]
- Cajal, S.R. El núcleo de las células piramidales del cerebro humano y de algunos mamíferos. Trab. Lab. Inv. Biol. Univ. Madr. 1910, 8, 27–62. [Google Scholar]
- Beck, J.S. Variations in the morphological patterns of "autoimmune" nuclear fluorescence. Lancet 1961, 1, 1203–1205. [Google Scholar] [CrossRef]
- Swift, H. Studies on nuclear fine structure. Brookhaven Symp. Biol. 1959, 12, 134–152. [Google Scholar]
- Perraud, M.; Gioud, M.; Monier, J.C. Intranuclear structures recognized by autoantibodies against ribonucleoproteins: Study on monkey kidney cells in culture using immunofluorescent techniques and immunoelectron microscopy. Ann. Immunol. (Paris) 1970, 130, 635–647. [Google Scholar]
- Lerner, E.A.; Lerner, M.R.; Janeway, C.A., Jr.; Steitz, J.A. Monoclonal antibodies to nucleic acid-containing cellular constituents: Probes for molecular biology and autoimmune disease. Proc. Natl. Acad. Sci. USA 1981, 78, 2737–2741. [Google Scholar] [CrossRef]
- Spector, D.L.; Schrier, W.H.; Busch, H. Immunoelectron microscopic localization of snRNPs. Biol. Cell 1983, 49, 1–10. [Google Scholar] [CrossRef]
- Girard, C.; Will, C.L.; Peng, J.; Makarov, E.M.; Kastner, B.; Lemm, I.; Urlaub, H.; Hartmuth, K.; Luhrmann, R. Post-transcriptional spliceosomes are retained in nuclear speckles until splicing completion. Nat. Commun. 2012, 3, 994. [Google Scholar]
- Lin, S.; Coutinho-Mansfield, G.; Wang, D.; Pandit, S.; Fu, X.D. The splicing factor SC35 has an active role in transcriptional elongation. Nat. Struct. Mol. Biol. 2008, 15, 819–826. [Google Scholar]
- Zhong, X.Y.; Wang, P.; Han, J.; Rosenfeld, M.G.; Fu, X.D. SR proteins in vertical integration of gene expression from transcription to RNA processing to translation. Mol. Cell 2009, 35, 1–10. [Google Scholar] [CrossRef]
- Chang, Y.F.; Imam, J.S.; Wilkinson, M.F. The nonsense-mediated decay RNA surveillance pathway. Annu. Rev. Biochem. 2007, 76, 51–74. [Google Scholar] [CrossRef]
- Wei, X.; Somanathan, S.; Samarabandu, J.; Berezney, R. Three-dimensional visualization of transcription sites and their association with splicing factor-rich nuclear speckles. J. Cell Biol. 1999, 146, 543–558. [Google Scholar] [CrossRef]
- Long, J.C.; Caceres, J.F. The SR protein family of splicing factors: Master regulators of gene expression. Biochem. J. 2009, 417, 15–27. [Google Scholar] [CrossRef]
- Misteli, T.; Caceres, J.F.; Clement, J.Q.; Krainer, A.R.; Wilkinson, M.F.; Spector, D.L. Serine phosphorylation of SR proteins is required for their recruitment to sites of transcription in vivo. J. Cell Biol. 1998, 143, 297–307. [Google Scholar]
- Strasser, K.; Masuda, S.; Mason, P.; Pfannstiel, J.; Oppizzi, M.; Rodriguez-Navarro, S.; Rondon, A.G.; Aguilera, A.; Struhl, K.; Reed, R.; et al. TREX is a conserved complex coupling transcription with messenger RNA export. Nature 2002, 417, 304–308. [Google Scholar] [CrossRef]
- Cmarko, D.; Verschure, P.J.; Martin, T.E.; Dahmus, M.E.; Krause, S.; Fu, X.D.; van Driel, R.; Fakan, S. Ultrastructural analysis of transcription and splicing in the cell nucleus after bromo-UTP microinjection. Mol. Biol. Cell 1999, 10, 211–223. [Google Scholar] [CrossRef]
- Jolly, C.; Usson, Y.; Morimoto, R.I. Rapid and reversible relocalization of heat shock factor 1 within seconds to nuclear stress granules. Proc. Natl. Acad. Sci. USA 1999, 96, 6769–6774. [Google Scholar] [CrossRef]
- Cotto, J.; Fox, S.; Morimoto, R. HSF1 granules: A novel stress-induced nuclear compartment of human cells. J. Cell Sci. 1997, 110, 2925–2934. [Google Scholar]
- Denegri, M.; Moralli, D.; Rocchi, M.; Biggiogera, M.; Raimondi, E.; Cobianchi, F.; De Carli, L.; Riva, S.; Biamonti, G. Human chromosomes 9, 12, and 15 contain the nucleation sites of stress-induced nuclear bodies. Mol. Biol. Cell 2002, 13, 2069–2079. [Google Scholar] [CrossRef]
- Morton, E.A.; Lamitina, T. Caenorhabditis elegans HSF-1 is an essential nuclear protein that forms stress granule-like structures following heat shock. Aging Cell 2012. [Google Scholar]
- Prasanth, K.V.; Rajendra, T.K.; Lal, A.K.; Lakhotia, S.C. Omega speckles—a novel class of nuclear speckles containing hnRNPs associated with noncoding hsr-omega RNA in Drosophila. J. Cell Sci. 2000, 113, 3485–3497. [Google Scholar]
- Mahl, P.; Lutz, Y.; Puvion, E.; Fuchs, J.P. Rapid effect of heat shock on two heterogeneous nuclear ribonucleoprotein-associated antigens in HeLa cells. J. Cell Biol. 1989, 109, 1921–1935. [Google Scholar] [CrossRef]
- Xiao, H.; Lis, J.T. Germline transformation used to define key features of heat-shock response elements. Science 1988, 239, 1139–1142. [Google Scholar]
- Jolly, C.; Konecny, L.; Grady, D.L.; Kutskova, Y.A.; Cotto, J.J.; Morimoto, R.I.; Vourc'h, C. In vivo binding of active heat shock transcription factor 1 to human chromosome 9 heterochromatin during stress. J. Cell Biol. 2002, 156, 775–781. [Google Scholar] [CrossRef]
- Oesterreich, S.; Lee, A.V.; Sullivan, T.M.; Samuel, S.K.; Davie, J.R.; Fuqua, S.A. Novel nuclear matrix protein HET binds to and influences activity of the HSP27 promoter in human breast cancer cells. J. Cell. Biochem. 1997, 67, 275–286. [Google Scholar] [CrossRef]
- Townson, S.M.; Kang, K.; Lee, A.V.; Oesterreich, S. Structure-function analysis of the estrogen receptor alpha corepressor scaffold attachment factor-B1: identification of a potent transcriptional repression domain. J. Biol. Chem. 2004, 279, 26074–26081. [Google Scholar]
- Montgomery, T.H. Comparative cytological studies, with especial regard to the morphology of the nucleolus. J. Morphol. 1898, 15, 265–582. [Google Scholar]
- Islinger, M.; Willimski, D.; Volkl, A.; Braunbeck, T. Effects of 17a-ethinylestradiol on the expression of three estrogen-responsive genes and cellular ultrastructure of liver and testes in male zebrafish. Aquat. Toxicol. 2003, 62, 85–103. [Google Scholar] [CrossRef]
- Knibiehler, B.; Mirre, C.; Rosset, R. Nucleolar organizer structure and activity in a nucleolus without fibrillar centres: The nucleolus in an established Drosophila cell line. J. Cell Sci. 1982, 57, 351–364. [Google Scholar]
- Lee, L.W.; Lee, C.C.; Huang, C.R.; Lo, S.J. The nucleolus of Caenorhabditis elegans. J. Biomed. Biotechnol. 2012, 2012, 601274. [Google Scholar]
- Smitt, W.W.; Vlak, J.M.; Molenaar, I.; Rozijn, T.H. Nucleolar function of the dense crescent in the yeast nucleus. A biochemical and ultrastructural study. Exp. Cell Res. 1973, 80, 313–321. [Google Scholar]
- Monneron, A.; Bernhard, W. Fine structural organization of the interphase nucleus in some mammalian cells. J. Ultrastruct. Res. 1969, 27, 266–288. [Google Scholar] [CrossRef]
- Wu, Z.A.; Murphy, C.; Callan, H.G.; Gall, J.G. Small nuclear ribonucleoproteins and heterogeneous nuclear ribonucleoproteins in the amphibian germinal vesicle: loops, spheres, and snurposomes. J. Cell Biol. 1991, 113, 465–483. [Google Scholar] [CrossRef]
- Segalat, L.; Lepesant, J.A. Spatial distribution of the Sm antigen in Drosophila early embryos. Biol. Cell 1992, 75, 181–185. [Google Scholar]
- Potashkin, J.A.; Derby, R.J.; Spector, D.L. Differential distribution of factors involved in pre-mRNA processing in the yeast cell nucleus. Mol. Cell. Biol. 1990, 10, 3524–3534. [Google Scholar]
- Schoenfelder, S.; Sexton, T.; Chakalova, L.; Cope, N.F.; Horton, A.; Andrews, S.; Kurukuti, S.; Mitchell, J.A.; Umlauf, D.; Dimitrova, D.S.; et al. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nat. Genet. 2010, 42, 53–61. [Google Scholar]
- Cajal, S.R. Un sencillo método de coloración seletiva del retículo protoplasmático y sus efectos en los diversos órganos nerviosos de vertebrados e invertebrados. Trab. Lab. Inv. Biol. Univ. Madr. 1903, 2, 129–221. [Google Scholar]
- Nizami, Z.F.; Gall, J.G. Pearls are novel Cajal body-like structures in the Xenopus germinal vesicle that are dependent on RNA pol III transcription. Chromosome Res. 2012, 20, 953–969. [Google Scholar] [CrossRef]
- Strzelecka, M.; Oates, A.C.; Neugebauer, K.M. Dynamic control of Cajal body number during zebrafish embryogenesis. Nucleus 2010, 1, 96–108. [Google Scholar]
- Verheggen, C.; Mouaikel, J.; Thiry, M.; Blanchard, J.M.; Tollervey, D.; Bordonne, R.; Lafontaine, D.L.; Bertrand, E. Box C/D small nucleolar RNA trafficking involves small nucleolar RNP proteins, nucleolar factors and a novel nuclear domain. EMBO J. 2001, 20, 5480–5490. [Google Scholar] [CrossRef]
- Francis, J.W.; Sandrock, A.W.; Bhide, P.G.; Vonsattel, J.P.; Brown, R.H., Jr. Heterogeneity of subcellular localization and electrophoretic mobility of survival motor neuron (SMN) protein in mammalian neural cells and tissues. Proc. Natl. Acad. Sci. USA 1998, 95, 6492–6497. [Google Scholar]
- Cauchi, R.J. Gem formation upon constitutive Gemin3 overexpression in Drosophila. Cell Biol. Int. 2011, 35, 1233–1238. [Google Scholar] [CrossRef]
- Wu, C.H.; Gall, J.G. U7 small nuclear RNA in C snurposomes of the Xenopus germinal vesicle. Proc. Natl. Acad. Sci. USA 1993, 90, 6257–6259. [Google Scholar] [CrossRef]
- Bongiorno-Borbone, L.; De Cola, A.; Vernole, P.; Finos, L.; Barcaroli, D.; Knight, R.A.; Melino, G.; De Laurenzi, V. FLASH and NPAT positive but not Coilin positive Cajal Bodies correlate with cell ploidy. Cell Cycle 2008, 7, 2357–2367. [Google Scholar]
- Ge, H.; Manley, J.L. A protein factor, ASF, controls cell-specific alternative splicing of SV40 early pre-mRNA in vitro. Cell 1990, 62, 25–34. [Google Scholar]
- Wu, H.; Sun, S.; Tu, K.; Gao, Y.; Xie, B.; Krainer, A.R.; Zhu, J. A splicing-independent function of SF2/ASF in microRNA processing. Mol. Cell 2010, 38, 67–77. [Google Scholar] [CrossRef]
- Jolly, C.; Vourc'h, C.; Robert-Nicoud, M.; Morimoto, R.I. Intron-independent association of splicing factors with active genes. J. Cell Biol. 1999, 145, 1133–1143. [Google Scholar] [CrossRef]
- Fu, X.D. Specific commitment of different pre-mRNAs to splicing by single SR proteins. Nature 1993, 365, 82–85. [Google Scholar]
- Busa, R.; Geremia, R.; Sette, C. Genotoxic stress causes the accumulation of the splicing regulator Sam68 in nuclear foci of transcriptionally active chromatin. Nucleic Acids Res. 2010, 38, 3005–3018. [Google Scholar] [CrossRef]
- Metz, A.; Soret, J.; Vourc'h, C.; Tazi, J.; Jolly, C. A key role for stress-induced satellite III transcripts in the relocalization of splicing factors into nuclear stress granules. J. Cell Sci. 2004, 117, 4551–4558. [Google Scholar]
- Shevtsov, S.P.; Dundr, M. Nucleation of nuclear bodies by RNA. Nat. Cell Biol. 2011, 13, 167–173. [Google Scholar]
- Fritah, S.; Col, E.; Boyault, C.; Govin, J.; Sadoul, K.; Chiocca, S.; Christians, E.; Khochbin, S.; Jolly, C.; Vourc'h, C. Heat-shock factor 1 controls genome-wide acetylation in heat-shocked cells. Mol. Biol. Cell 2009, 20, 4976–4984. [Google Scholar]
- Papantonis, A.; Larkin, J.D.; Wada, Y.; Ohta, Y.; Ihara, S.; Kodama, T.; Cook, P.R. Active RNA polymerases: Mobile or immobile molecular machines? PLoS Biol. 2010, 8, e1000419. [Google Scholar] [CrossRef]
- Dickinson, P.; Cook, P.R.; Jackson, D.A. Active RNA polymerase I is fixed within the nucleus of HeLa cells. EMBO J. 1990, 9, 2207–2214. [Google Scholar]
- Mitchell, J.A.; Fraser, P. Transcription factories are nuclear subcompartments that remain in the absence of transcription. Genes Dev. 2008, 22, 20–25. [Google Scholar] [CrossRef]
- Cook, P.R. Predicting three-dimensional genome structure from transcriptional activity. Nat. Genet. 2002, 32, 347–352. [Google Scholar] [CrossRef]
- McCracken, S.; Fong, N.; Yankulov, K.; Ballantyne, S.; Pan, G.; Greenblatt, J.; Patterson, S.D.; Wickens, M.; Bentley, D.L. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature 1997, 385, 357–361. [Google Scholar] [CrossRef]
- Gall, J.G. The centennial of the Cajal body. Nat. Rev. Mol. Cell Biol. 2003, 4, 975–980. [Google Scholar] [CrossRef]
- Dundr, M. Nuclear bodies: Multifunctional companions of the genome. Curr. Opin. Cell Biol. 2012, 24, 415–422. [Google Scholar] [CrossRef]
- Machyna, M.; Heyn, P.; Neugebauer, K.M. Cajal bodies: Where form meets function. Wiley Interdiscip Rev RNA 2013, 4, 17–34. [Google Scholar] [CrossRef]
- Jady, B.E.; Bertrand, E.; Kiss, T. Human telomerase RNA and box H/ACA scaRNAs share a common Cajal body-specific localization signal. J. Cell Biol. 2004, 164, 647–652. [Google Scholar] [CrossRef]
- Baillat, D.; Hakimi, M.A.; Naar, A.M.; Shilatifard, A.; Cooch, N.; Shiekhattar, R. Integrator, a multiprotein mediator of small nuclear RNA processing, associates with the C-terminal repeat of RNA polymerase II. Cell 2005, 123, 265–276. [Google Scholar] [CrossRef]
- Takata, H.; Nishijima, H.; Maeshima, K.; Shibahara, K. The integrator complex is required for integrity of Cajal bodies. J. Cell Sci. 2012, 125, 166–175. [Google Scholar] [CrossRef]
- Xu, H.; Pillai, R.S.; Azzouz, T.N.; Shpargel, K.B.; Kambach, C.; Hebert, M.D.; Schumperli, D.; Matera, A.G. The C-terminal domain of coilin interacts with Sm proteins and U snRNPs. Chromosoma 2005, 114, 155–166. [Google Scholar] [CrossRef]
- Broome, H.J.; Hebert, M.D. Coilin displays differential affinity for specific RNAs in vivo and is linked to telomerase RNA biogenesis. J. Mol. Biol. 2013. [Google Scholar]
- Hebert, M.D.; Szymczyk, P.W.; Shpargel, K.B.; Matera, A.G. Coilin forms the bridge between Cajal bodies and SMN, the spinal muscular atrophy protein. Genes Dev. 2001, 15, 2720–2729. [Google Scholar] [CrossRef]
- Broome, H.J.; Hebert, M.D. In vitro RNase and nucleic acid binding activities implicate coilin in U snRNA processing. PLoS One 2012, 7, e36300. [Google Scholar] [CrossRef]
- Sleeman, J.E.; Lamond, A.I. Newly assembled snRNPs associate with coiled bodies before speckles, suggesting a nuclear snRNP maturation pathway. Curr. Biol. 1999, 9, 1065–1074. [Google Scholar] [CrossRef]
- Stanek, D.; Neugebauer, K.M. Detection of snRNP assembly intermediates in Cajal bodies by fluorescence resonance energy transfer. J. Cell Biol. 2004, 166, 1015–1025. [Google Scholar] [CrossRef]
- Nesic, D.; Tanackovic, G.; Kramer, A. A role for Cajal bodies in the final steps of U2 snRNP biogenesis. J. Cell Sci. 2004, 117, 4423–4433. [Google Scholar] [CrossRef]
- Kiss, T.; Fayet, E.; Jady, B.E.; Richard, P.; Weber, M. Biogenesis and intranuclear trafficking of human box C/D and H/ACA RNPs. Cold Spring Harb. Symp. Quant. Biol. 2006, 71, 407–417. [Google Scholar] [CrossRef]
- Mahmoudi, S.; Henriksson, S.; Weibrecht, I.; Smith, S.; Soderberg, O.; Stromblad, S.; Wiman, K.G.; Farnebo, M. WRAP53 is essential for Cajal body formation and for targeting the survival of motor neuron complex to Cajal bodies. PLoS Biol. 2010, 8, e1000521. [Google Scholar] [CrossRef]
- Davis, D.R. Stabilization of RNA stacking by pseudouridine. Nucleic Acids Res. 1995, 23, 5020–5026. [Google Scholar] [CrossRef]
- Wu, G.; Xiao, M.; Yang, C.; Yu, Y.T. U2 snRNA is inducibly pseudouridylated at novel sites by Pus7p and snR81 RNP. EMBO J. 2011, 30, 79–89. [Google Scholar] [CrossRef]
- Frey, M.R.; Bailey, A.D.; Weiner, A.M.; Matera, A.G. Association of snRNA genes with coiled bodies is mediated by nascent snRNA transcripts. Curr. Biol. 1999, 9, 126–135. [Google Scholar] [CrossRef]
- Smith, K.P.; Carter, K.C.; Johnson, C.V.; Lawrence, J.B. U2 and U1 snRNA gene loci associate with coiled bodies. J. Cell. Biochem. 1995, 59, 473–485. [Google Scholar]
- Smith, K.P.; Lawrence, J.B. Interactions of U2 gene loci and their nuclear transcripts with Cajal (coiled) bodies: Evidence for PreU2 within Cajal bodies. Mol. Biol. Cell 2000, 11, 2987–2998. [Google Scholar] [CrossRef]
- Sleeman, J.E.; Lamond, A.I. Newly assembled snRNPs associate with coiled bodies before speckles, suggesting a nuclear snRNP maturation pathway. Curr. Biol. 1999, 9, 1065–1074. [Google Scholar] [CrossRef]
- Cristofari, G.; Adolf, E.; Reichenbach, P.; Sikora, K.; Terns, R.M.; Terns, M.P.; Lingner, J. Human telomerase RNA accumulation in Cajal bodies facilitates telomerase recruitment to telomeres and telomere elongation. Mol. Cell 2007, 27, 882–889. [Google Scholar]
- Tomlinson, R.L.; Abreu, E.B.; Ziegler, T.; Ly, H.; Counter, C.M.; Terns, R.M.; Terns, M.P. Telomerase reverse transcriptase is required for the localization of telomerase RNA to Cajal bodies and telomeres in human cancer cells. Mol. Biol. Cell 2008, 19, 3793–3800. [Google Scholar] [CrossRef]
- Tomlinson, R.L.; Ziegler, T.D.; Supakorndej, T.; Terns, R.M.; Terns, M.P. Cell cycle-regulated trafficking of human telomerase to telomeres. Mol. Biol. Cell 2006, 17, 955–965. [Google Scholar]
- Jady, B.E.; Richard, P.; Bertrand, E.; Kiss, T. Cell cycle-dependent recruitment of telomerase RNA and Cajal bodies to human telomeres. Mol. Biol. Cell 2006, 17, 944–954. [Google Scholar]
- Werdnig, G. Zwei frühinfantile hereditäre fälle von progressiver muskelatrophie unter dem bilde der dystrophie, aber auf neurotischer grundlage. Arch. Psychiatr. Nervenkr. 1891, 22, 437–481. [Google Scholar] [CrossRef]
- Hoffmann, J. Weitere beiträge zur lehre von der progressiven neurotischen muskeldystrophie. Dtsch. Z. Nervenheilkd. 1891, 1, 95–120. [Google Scholar] [CrossRef]
- Strasswimmer, J.; Lorson, C.L.; Breiding, D.E.; Chen, J.J.; Le, T.; Burghes, A.H.; Androphy, E.J. Identification of survival motor neuron as a transcriptional activator-binding protein. Hum. Mol. Genet. 1999, 8, 1219–1226. [Google Scholar]
- Pellizzoni, L.; Charroux, B.; Rappsilber, J.; Mann, M.; Dreyfuss, G. A functional interaction between the survival motor neuron complex and RNA polymerase II. J. Cell Biol. 2001, 152, 75–85. [Google Scholar]
- Sanchez, G.; Dury, A.Y.; Murray, L.M.; Biondi, O.; Tadesse, H.; El Fatimy, R.; Kothary, R.; Charbonnier, F.; Khandjian, E.W.; Cote, J. A novel function for the survival motoneuron protein as a translational regulator. Hum. Mol. Genet. 2012. [Google Scholar]
- Battle, D.J.; Lau, C.K.; Wan, L.; Deng, H.; Lotti, F.; Dreyfuss, G. The Gemin5 protein of the SMN complex identifies snRNAs. Mol. Cell 2006, 23, 273–279. [Google Scholar]
- Galcheva-Gargova, Z.; Gangwani, L.; Konstantinov, K.N.; Mikrut, M.; Theroux, S.J.; Enoch, T.; Davis, R.J. The cytoplasmic zinc finger protein ZPR1 accumulates in the nucleolus of proliferating cells. Mol. Biol. Cell 1998, 9, 2963–2971. [Google Scholar] [CrossRef]
- Galcheva-Gargova, Z.; Konstantinov, K.N.; Wu, I.H.; Klier, F.G.; Barrett, T.; Davis, R.J. Binding of zinc finger protein ZPR1 to the epidermal growth factor receptor. Science 1996, 272, 1797–1802. [Google Scholar]
- Hebert, M.D.; Shpargel, K.B.; Ospina, J.K.; Tucker, K.E.; Matera, A.G. Coilin methylation regulates nuclear body formation. Dev. Cell 2002, 3, 329–337. [Google Scholar] [CrossRef]
- Shafey, D.; Cote, P.D.; Kothary, R. Hypomorphic Smn knockdown C2C12 myoblasts reveal intrinsic defects in myoblast fusion and myotube morphology. Exp. Cell Res. 2005, 311, 49–61. [Google Scholar] [CrossRef]
- Yang, X.C.; Sabath, I.; Debski, J.; Kaus-Drobek, M.; Dadlez, M.; Marzluff, W.F.; Dominski, Z. A complex containing the CPSF73 endonuclease and other polyadenylation factors associates with U7 snRNP and is recruited to histone pre-mRNA for 3'-end processing. Mol. Cell. Biol. 2013, 33, 28–37. [Google Scholar] [CrossRef]
- Sanchez, R.; Marzluff, W.F. The stem-loop binding protein is required for efficient translation of histone mRNA in vivo and in vitro. Mol. Cell. Biol. 2002, 22, 7093–7104. [Google Scholar] [CrossRef]
- Krishnan, N.; Lam, T.T.; Fritz, A.; Rempinski, D.; O'Loughlin, K.; Minderman, H.; Berezney, R.; Marzluff, W.F.; Thapar, R. The prolyl isomerase Pin1 targets stem-loop binding protein (SLBP) to dissociate the SLBP-histone mRNA complex linking histone mRNA decay with SLBP ubiquitination. Mol. Cell. Biol. 2012, 32, 4306–4322. [Google Scholar] [CrossRef]
- Fox, A.H.; Bond, C.S.; Lamond, A.I. p54nrb forms a heterodimer with PSP1 that localizes to paraspeckles in an RNA-dependent manner. Mol. Biol. Cell 2005, 16, 5304–5315. [Google Scholar] [CrossRef]
- Naganuma, T.; Nakagawa, S.; Tanigawa, A.; Sasaki, Y.F.; Goshima, N.; Hirose, T. Alternative 3'-end processing of long noncoding RNA initiates construction of nuclear paraspeckles. EMBO J. 2012, 31, 4020–4034. [Google Scholar] [CrossRef]
- Rosonina, E.; Ip, J.Y.; Calarco, J.A.; Bakowski, M.A.; Emili, A.; McCracken, S.; Tucker, P.; Ingles, C.J.; Blencowe, B.J. Role for PSF in mediating transcriptional activator-dependent stimulation of pre-mRNA processing in vivo. Mol. Cell. Biol. 2005, 25, 6734–6746. [Google Scholar] [CrossRef]
- Kaneko, S.; Rozenblatt-Rosen, O.; Meyerson, M.; Manley, J.L. The multifunctional protein p54nrb/PSF recruits the exonuclease XRN2 to facilitate pre-mRNA 3' processing and transcription termination. Genes Dev. 2007, 21, 1779–1789. [Google Scholar] [CrossRef]
- Patton, J.G.; Porro, E.B.; Galceran, J.; Tempst, P.; Nadal-Ginard, B. Cloning and characterization of PSF, a novel pre-mRNA splicing factor. Genes Dev. 1993, 7, 393–406. [Google Scholar] [CrossRef]
- Peng, R.; Dye, B.T.; Perez, I.; Barnard, D.C.; Thompson, A.B.; Patton, J.G. PSF and p54nrb bind a conserved stem in U5 snRNA. RNA 2002, 8, 1334–1347. [Google Scholar] [CrossRef]
- Kameoka, S.; Duque, P.; Konarska, M.M. p54(nrb) associates with the 5' splice site within large transcription/splicing complexes. EMBO J. 2004, 23, 1782–1791. [Google Scholar] [CrossRef]
- Bladen, C.L.; Udayakumar, D.; Takeda, Y.; Dynan, W.S. Identification of the polypyrimidine tract binding protein-associated splicing factor p54(nrb) complex as a candidate DNA double-strand break rejoining factor. J. Biol. Chem. 2005, 280, 5205–5210. [Google Scholar]
- Li, S.; Kuhne, W.W.; Kulharya, A.; Hudson, F.Z.; Ha, K.; Cao, Z.; Dynan, W.S. Involvement of p54(nrb), a PSF partner protein, in DNA double-strand break repair and radioresistance. Nucleic Acids Res. 2009, 37, 6746–6753. [Google Scholar] [CrossRef]
- Salton, M.; Lerenthal, Y.; Wang, S.Y.; Chen, D.J.; Shiloh, Y. Involvement of Matrin 3 and SFPQ/NONO in the DNA damage response. Cell Cycle 2010, 9, 1568–1576. [Google Scholar] [CrossRef]
- Ha, K.; Takeda, Y.; Dynan, W.S. Sequences in PSF/SFPQ mediate radioresistance and recruitment of PSF/SFPQ-containing complexes to DNA damage sites in human cells. DNA Repair (Amst) 2011, 10, 252–259. [Google Scholar] [CrossRef]
- Lowery, L.A.; Rubin, J.; Sive, H. Whitesnake/sfpq is required for cell survival and neuronal development in the zebrafish. Dev. Dyn. 2007, 236, 1347–1357. [Google Scholar]
- Auboeuf, D.; Dowhan, D.H.; Li, X.; Larkin, K.; Ko, L.; Berget, S.M.; O'Malley, B.W. CoAA, a nuclear receptor coactivator protein at the interface of transcriptional coactivation and RNA splicing. Mol. Cell. Biol. 2004, 24, 442–453. [Google Scholar] [CrossRef]
- Ruegsegger, U.; Beyer, K.; Keller, W. Purification and characterization of human cleavage factor Im involved in the 3' end processing of messenger RNA precursors. J. Biol. Chem. 1996, 271, 6107–6113. [Google Scholar] [CrossRef]
- Ruepp, M.D.; Aringhieri, C.; Vivarelli, S.; Cardinale, S.; Paro, S.; Schumperli, D.; Barabino, S.M. Mammalian pre-mRNA 3' end processing factor CF I m 68 functions in mRNA export. Mol. Biol. Cell 2009, 20, 5211–5223. [Google Scholar] [CrossRef]
- Zhang, Z.; Carmichael, G.G. The fate of dsRNA in the nucleus: A p54(nrb)-containing complex mediates the nuclear retention of promiscuously A-to-I edited RNAs. Cell 2001, 106, 465–475. [Google Scholar] [CrossRef]
- Moncada, S.; Higgs, A. The L-arginine-nitric oxide pathway. N. Engl. J. Med. 1993, 329, 2002–2012. [Google Scholar] [CrossRef]
- Herman, R.C.; Williams, J.G.; Penman, S. Message and non-message sequences adjacent to poly(A) in steady state heterogeneous nuclear RNA of HeLa cells. Cell 1976, 7, 429–437. [Google Scholar]
- Athanasiadis, A.; Rich, A.; Maas, S. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol. 2004, 2, e391. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.D.; Kim, T.T.; Walsh, T.; Kobayashi, Y.; Matise, T.C.; Buyske, S.; Gabriel, A. Widespread RNA editing of embedded alu elements in the human transcriptome. Genome Res. 2004, 14, 1719–1725. [Google Scholar] [CrossRef]
- Levanon, E.Y.; Eisenberg, E.; Yelin, R.; Nemzer, S.; Hallegger, M.; Shemesh, R.; Fligelman, Z.Y.; Shoshan, A.; Pollock, S.R.; Sztybel, D.; et al. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat. Biotechnol. 2004, 22, 1001–1005. [Google Scholar]
- Rabl, C. Über Zelltheilung. Morphol. Jahrbuch 1885, 10, 214–330. [Google Scholar]
- Bischoff, A.; Albers, J.; Kharboush, I.; Stelzer, E.; Cremer, T.; Cremer, C. Differences of size and shape of active and inactive X-chromosome domains in human amniotic fluid cell nuclei. Microsc. Res. Tech. 1993, 25, 68–77. [Google Scholar] [CrossRef]
- Cremer, T.; Cremer, C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat. Rev. Genet. 2001, 2, 292–301. [Google Scholar] [CrossRef]
- Cremer, T.; Lichter, P.; Borden, J.; Ward, D.C.; Manuelidis, L. Detection of chromosome aberrations in metaphase and interphase tumor cells by in situ hybridization using chromosome-specific library probes. Hum. Genet. 1988, 80, 235–246. [Google Scholar] [CrossRef]
- Lichter, P.; Cremer, T.; Borden, J.; Manuelidis, L.; Ward, D.C. Delineation of individual human chromosomes in metaphase and interphase cells by in situ suppression hybridization using recombinant DNA libraries. Hum. Genet. 1988, 80, 224–234. [Google Scholar] [CrossRef]
- Pinkel, D.; Landegent, J.; Collins, C.; Fuscoe, J.; Segraves, R.; Lucas, J.; Gray, J. Fluorescence in situ hybridization with human chromosome-specific libraries: detection of trisomy 21 and translocations of chromosome 4. Proc. Natl. Acad. Sci. USA 1988, 85, 9138–9142. [Google Scholar]
- Chambeyron, S.; Bickmore, W.A. Chromatin decondensation and nuclear reorganization of the HoxB locus upon induction of transcription. Genes Dev. 2004, 18, 1119–1130. [Google Scholar] [CrossRef]
- Chambeyron, S.; Da Silva, N.R.; Lawson, K.A.; Bickmore, W.A. Nuclear re-organisation of the Hoxb complex during mouse embryonic development. Development 2005, 132, 2215–2223. [Google Scholar] [CrossRef]
- Dietzel, S.; Schiebel, K.; Little, G.; Edelmann, P.; Rappold, G.A.; Eils, R.; Cremer, C.; Cremer, T. The 3D positioning of ANT2 and ANT3 genes within female X chromosome territories correlates with gene activity. Exp. Cell Res. 1999, 252, 363–375. [Google Scholar] [CrossRef]
- Ferrai, C.; Xie, S.Q.; Luraghi, P.; Munari, D.; Ramirez, F.; Branco, M.R.; Pombo, A.; Crippa, M.P. Poised transcription factories prime silent uPA gene prior to activation. PLoS Biol. 2010, 8, e1000270. [Google Scholar] [CrossRef]
- Kurz, A.; Lampel, S.; Nickolenko, J.E.; Bradl, J.; Benner, A.; Zirbel, R.M.; Cremer, T.; Lichter, P. Active and inactive genes localize preferentially in the periphery of chromosome territories. J. Cell Biol. 1996, 135, 1195–1205. [Google Scholar] [CrossRef]
- Mahy, N.L.; Perry, P.E.; Bickmore, W.A. Gene density and transcription influence the localization of chromatin outside of chromosome territories detectable by FISH. J. Cell Biol. 2002, 159, 753–763. [Google Scholar] [CrossRef]
- Mahy, N.L.; Perry, P.E.; Gilchrist, S.; Baldock, R.A.; Bickmore, W.A. Spatial organization of active and inactive genes and noncoding DNA within chromosome territories. J. Cell Biol. 2002, 157, 579–589. [Google Scholar] [CrossRef]
- Morey, C.; Da Silva, N.R.; Perry, P.; Bickmore, W.A. Nuclear reorganisation and chromatin decondensation are conserved, but distinct, mechanisms linked to Hox gene activation. Development 2007, 134, 909–919. [Google Scholar] [CrossRef]
- Scheuermann, M.O.; Tajbakhsh, J.; Kurz, A.; Saracoglu, K.; Eils, R.; Lichter, P. Topology of genes and nontranscribed sequences in human interphase nuclei. Exp. Cell Res. 2004, 301, 266–279. [Google Scholar] [CrossRef]
- Volpi, E.V.; Chevret, E.; Jones, T.; Vatcheva, R.; Williamson, J.; Beck, S.; Campbell, R.D.; Goldsworthy, M.; Powis, S.H.; Ragoussis, J.; et al. Large-scale chromatin organization of the major histocompatibility complex and other regions of human chromosome 6 and its response to interferon in interphase nuclei. J. Cell Sci. 2000, 113, 1565–1576. [Google Scholar]
- Williams, R.R.; Broad, S.; Sheer, D.; Ragoussis, J. Subchromosomal positioning of the epidermal differentiation complex (EDC) in keratinocyte and lymphoblast interphase nuclei. Exp. Cell Res. 2002, 272, 163–175. [Google Scholar] [CrossRef]
- Bridger, J.M.; Herrmann, H.; Munkel, C.; Lichter, P. Identification of an interchromosomal compartment by polymerization of nuclear-targeted vimentin. J. Cell Sci. 1998, 111, 1241–1253. [Google Scholar]
- Clemson, C.M.; Lawrence, J.B. Multifunctional compartments in the nucleus: Insights from DNA and RNA localization. J. Cell. Biochem. 1996, 62, 181–190. [Google Scholar] [CrossRef]
- Lampel, S.; Bridger, J.M.; Zirbel, R.M.; Mathieu, U.R.; Lichter, P. Nuclear RNA accumulations contain released transcripts and exhibit specific distributions with respect to Sm antigen foci. DNA Cell Biol. 1997, 16, 1133–1142. [Google Scholar] [CrossRef]
- Xing, Y.; Johnson, C.V.; Moen, P.T., Jr.; McNeil, J.A.; Lawrence, J. Nonrandom gene organization: Structural arrangements of specific pre-mRNA transcription and splicing with SC-35 domains. J. Cell Biol. 1995, 131, 1635–1647. [Google Scholar] [CrossRef]
- Zirbel, R.M.; Mathieu, U.R.; Kurz, A.; Cremer, T.; Lichter, P. Evidence for a nuclear compartment of transcription and splicing located at chromosome domain boundaries. Chromosome Res. 1993, 1, 93–106. [Google Scholar] [CrossRef]
- Bickmore, W.A.; Chubb, J.R. Dispatch. Chromosome position: Now, where was I? Curr. Biol. 2003, 13, R357–R359. [Google Scholar]
- Gerlich, D.; Beaudouin, J.; Kalbfuss, B.; Daigle, N.; Eils, R.; Ellenberg, J. Global chromosome positions are transmitted through mitosis in mammalian cells. Cell 2003, 112, 751–764. [Google Scholar] [CrossRef]
- Parada, L.A.; Roix, J.J.; Misteli, T. An uncertainty principle in chromosome positioning. Trends Cell Biol. 2003, 13, 393–396. [Google Scholar] [CrossRef]
- Walter, J.; Schermelleh, L.; Cremer, M.; Tashiro, S.; Cremer, T. Chromosome order in HeLa cells changes during mitosis and early G1, but is stably maintained during subsequent interphase stages. J. Cell Biol. 2003, 160, 685–697. [Google Scholar] [CrossRef]
- Williams, R.R.; Fisher, A.G. Chromosomes, positions please! Nat. Cell Biol. 2003, 5, 388–390. [Google Scholar] [CrossRef]
- Harewood, L.; Schutz, F.; Boyle, S.; Perry, P.; Delorenzi, M.; Bickmore, W.A.; Reymond, A. The effect of translocation-induced nuclear reorganization on gene expression. Genome Res. 2010, 20, 554–564. [Google Scholar] [CrossRef]
- Campalans, A.; Amouroux, R.; Bravard, A.; Epe, B.; Radicella, J.P. UVA irradiation induces relocalisation of the DNA repair protein hOGG1 to nuclear speckles. J. Cell Sci. 2007, 120, 23–32. [Google Scholar]
- Ishitani, K.; Yoshida, T.; Kitagawa, H.; Ohta, H.; Nozawa, S.; Kato, S. p54nrb acts as a transcriptional coactivator for activation function 1 of the human androgen receptor. Biochem. Biophys. Res. Commun. 2003, 306, 660–665. [Google Scholar] [CrossRef]
- Kuwahara, S.; Ikei, A.; Taguchi, Y.; Tabuchi, Y.; Fujimoto, N.; Obinata, M.; Uesugi, S.; Kurihara, Y. PSPC1, NONO, and SFPQ are expressed in mouse Sertoli cells and may function as coregulators of androgen receptor-mediated transcription. Biol. Reprod. 2006, 75, 352–359. [Google Scholar] [CrossRef]
- Mayer, C.; Bierhoff, H.; Grummt, I. The nucleolus as a stress sensor: JNK2 inactivates the transcription factor TIF-IA and down-regulates rRNA synthesis. Genes Dev. 2005, 19, 933–941. [Google Scholar] [CrossRef]
- Velma, V.; Carrero, Z.I.; Cosman, A.M.; Hebert, M.D. Coilin interacts with Ku proteins and inhibits in vitro non-homologous DNA end joining. FEBS Lett. 2010, 584, 4735–4739. [Google Scholar] [CrossRef]
- Xu, M.; Cook, P.R. The role of specialized transcription factories in chromosome pairing. Biochim. Biophys. Acta 2008, 1783, 2155–2160. [Google Scholar]
- Zhao, J.; Kennedy, B.K.; Lawrence, B.D.; Barbie, D.A.; Matera, A.G.; Fletcher, J.A.; Harlow, E. NPAT links cyclin E-Cdk2 to the regulation of replication-dependent histone gene transcription. Genes Dev. 2000, 14, 2283–2297. [Google Scholar] [CrossRef]
- Mao, Y.S.; Zhang, B.; Spector, D.L. Biogenesis and function of nuclear bodies. Trends Genet. 2011, 27, 295–306. [Google Scholar] [CrossRef]
- Dundr, M.; Misteli, T. Biogenesis of nuclear bodies. Cold Spring Harb. Perspect. Biol. 2010, 2, a000711. [Google Scholar] [CrossRef]
- Shav-Tal, Y.; Blechman, J.; Darzacq, X.; Montagna, C.; Dye, B.T.; Patton, J.G.; Singer, R.H.; Zipori, D. Dynamic sorting of nuclear components into distinct nucleolar caps during transcriptional inhibition. Mol. Biol. Cell 2005, 16, 2395–2413. [Google Scholar] [CrossRef]
- Spector, D.L.; Fu, X.D.; Maniatis, T. Associations between distinct pre-mRNA splicing components and the cell nucleus. EMBO J. 1991, 10, 3467–3481. [Google Scholar]
- Young, P.J.; Le, T.T.; Dunckley, M.; Nguyen, T.M.; Burghes, A.H.; Morris, G.E. Nuclear gems and Cajal (coiled) bodies in fetal tissues: nucleolar distribution of the spinal muscular atrophy protein, SMN. Exp. Cell Res. 2001, 265, 252–261. [Google Scholar] [CrossRef]
- Gdula, M.R.; Poterlowicz, K.; Mardaryev, A.N.; Sharov, A.A.; Peng, Y.; Fessing, M.Y.; Botchkarev, V.A. Remodeling of Three-Dimensional Organization of the Nucleus during Terminal Keratinocyte Differentiation in the Epidermis. J. Invest. Dermatol. 2013. [Google Scholar]
- Berrios, S.; Fernandez-Donoso, R.; Pincheira, J.; Page, J.; Manterola, M.; Cerda, M.C. Number and nuclear localisation of nucleoli in mammalian spermatocytes. Genetica 2004, 121, 219–228. [Google Scholar] [CrossRef]
- McClintock, B. The relation of a particular chromosomal element to the development of the nucleoli in Zea mays. Z. Zellforsch. 1934, 21, 294–328. [Google Scholar] [CrossRef]
- Pardue, M.L.; Hsu, T.C. Locations of 18S and 28S ribosomal genes on the chromosomes of the Indian muntjac. J. Cell Biol. 1975, 64, 251–254. [Google Scholar] [CrossRef]
- Lima-de-Faria, A. The relation between chromomeres, replicons, operons, transcription units, genes, viruses and palindromes. Hereditas 1975, 81, 249–284. [Google Scholar] [CrossRef]
- Lima-de-Faria, A. Classification of genes, rearrangements and chromosomes according to the chromosome field. Hereditas 1980, 93, 1–46. [Google Scholar] [CrossRef]
- Thatcher, T.H.; Gorovsky, M.A. Phylogenetic analysis of the core histones H2A, H2B, H3, and H4. Nucleic Acids Res. 1994, 22, 174–179. [Google Scholar] [CrossRef]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Morimoto, M.; Boerkoel, C.F. The Role of Nuclear Bodies in Gene Expression and Disease. Biology 2013, 2, 976-1033. https://doi.org/10.3390/biology2030976
Morimoto M, Boerkoel CF. The Role of Nuclear Bodies in Gene Expression and Disease. Biology. 2013; 2(3):976-1033. https://doi.org/10.3390/biology2030976
Chicago/Turabian StyleMorimoto, Marie, and Cornelius F. Boerkoel. 2013. "The Role of Nuclear Bodies in Gene Expression and Disease" Biology 2, no. 3: 976-1033. https://doi.org/10.3390/biology2030976
APA StyleMorimoto, M., & Boerkoel, C. F. (2013). The Role of Nuclear Bodies in Gene Expression and Disease. Biology, 2(3), 976-1033. https://doi.org/10.3390/biology2030976