Eukaryotic LYR Proteins Interact with Mitochondrial Protein Complexes
Abstract
:1. The LYR Motif
| Human Protein | m (kDa) | pI | Mitochondrial Localization | Import Probability [21] | Section of Sequence Alignment a (Numbers Indicate Amino Acid Residues) [22] | Function | Interaction Partner |
|---|---|---|---|---|---|---|---|
| LYRM1 | 14 | 10 | Yes [3] | 93 |  | insulin signaling | unknown |
| LYRM2 | 11 | 11 | Yes [3] | 99 |  | unknown | unknown |
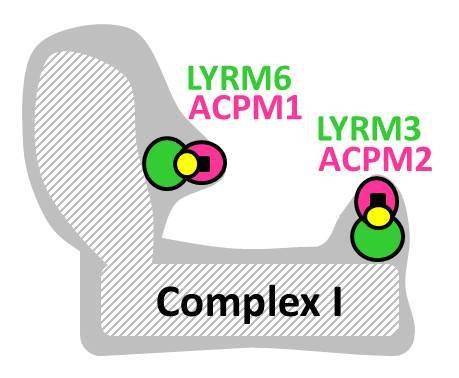
| LYRM3/NDUFB9 | 22 | 9 | Yes [3] | 90 |  | CI subunit | ACPM |
| LYRM4/ISD11 | 11 | 11 | Yes [3] | 92 |  | ISC biogenesis | NFS1, ACPM |
| LYRM5 | 11 | 10 | Yes [3] | 35 |  | COPP | unknown |
| LYRM6/NDUFA6 | 18 | 10 | Yes [3] | 94 |  | CI subunit | NDUFS3, ACPM |
| LYRM7/MZM1L | 12 | 10 | Yes [3] | 87 |  | CIII AF | UQCRFS1, HSC20 |
| LYRM8/SDHAF1, yeast Sdh6 | 13 | 11 | Yes [14] | 29 |  | CII AF | SDHB, HSC20 |
| LYRM9 | 10 | 9 | unknown | 99 |  | unknown | unknown |
| ACN9/SDHAF3, yeast Sdh7 | 15 | 9 | Yes [4,5] | 98 |  | CII AF | SDHB |
| FMC1/C7orf55 b | 13 | 10 | Yes [6] | 90 |  | CV AF | ATP12, ACPM |


2. The Diverse Roles of the LYR Motif Proteins in Mitochondria
2.1. Complex I Subunits LYRM3, LYRM6 and ACPM
2.2. Essential Factor of Fe-S Cluster Biogenesis (LYRM4)

2.3. Complex I-Related LYRM5
2.4. Assembly Factors of OXPHOS Complexes (LYRM7, LYRM8, ACN9, FMC1)
2.5. LYRM1, LYRM2 and LYRM9
3. Multi-Domain Organization of LYRMs
4. Conclusions
| LYRM a | Fused Domain | Organism(s) | Source | LYRM Domain, Amino Acid Residue (Start–End) | Human Orthologue | Pfam Domain, Remarks |
|---|---|---|---|---|---|---|
| LYRM1 | Ras | Salpingoeca rosetta | http://pfam.xfam.org/protein/F2USF2_SALS5 | 180–230 | RAB8B_HUMAN b | PF00071, small GTP binding protein RAB8 |
| LYRM2 | BTB-SPRY | Naegleria gruberi | http://pfam.xfam.org/protein/D2VKV5_NAEGR | 462–524 | KBTB3_HUMAN | PF00651/PF00622, BTB/POZ protein |
| LYRM3 | DEAD/DEAH | Selaginella moellendorffii | http://pfam.xfam.org/protein/D8TAY1_SELML | 22–81 | DDX28_HUMAN b | PF00270, RNA helicase, RNA processing |
| LYRM4 | RF-1 | Drosophila persimili c | http://pfam.xfam.org/protein/B4GAC8_DROPE | 7–61 | CL065_HUMAN b | PF00472, translational release factor 1, C12orf65 |
| LYRM7 | CS (HSP20-like) | Dictyostelium fasciculatum | http://pfam.xfam.org/protein/F4Q9B8_DICFS | 6–68 | ─ | PF04969, binding module for HSP90 |
| LYRM8 | HSP20 | Penicillium marneffei d | http://pfam.xfam.org/protein/B6QRH6_PENMQ | 105–164 | CRYAA_HUMAN | PF00011 |
| LYRM9 | NO_synthase | Cricetulus griseus | http://pfam.xfam.org/protein/G3GTQ5_CRIGR | 11–69 | NOS2_HUMAN | PF02898, inducible NO synthase |
| ACN9 | RRP7 | Pichia pastoris | http://pfam.xfam.org/protein/F2QP71_PICP7 | 10–67 | RRP7A_HUMAN | PF12923, ribosomal RNA-processing protein 7 |
| FMC1 | Ras-GEF | Talaromyces stipitatus e | http://pfam.xfam.org/protein/B8MGM3_TALSN | 568–659 | RPGF3_HUMAN | PF00617, RAP guanine nucleotide exchange factor 3 |
Supplementary Files
Supplementary File 1Acknowledgments
Conflicts of Interest
References
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J.; et al. Pfam: The protein families database. Nucleic Acids Res. 2014, 42, D222–D230. [Google Scholar] [CrossRef]
- Angerer, H. The superfamily of mitochondrial Complex1_LYR motif-containing (LYRM) proteins. Biochem. Soc. Trans. 2013, 41, 1335–1341. [Google Scholar] [CrossRef] [PubMed]
- Pagliarini, D.J.; Calvo, S.E.; Chang, B.; Sheth, S.A.; Vafai, S.B.; Ong, S.E.; Walford, G.A.; Sugiana, C.; Boneh, A.; Chen, W.K.; et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell 2008, 134, 112–123. [Google Scholar]
- Dennis, R.A.; McCammon, M.T. ACN9 is a novel protein of gluconeogenesis that is located in the mitochondrial intermembrane space. Eur. J. Biochem. 1999, 261, 236–243. [Google Scholar] [CrossRef] [PubMed]
- Na, U.; Yu, W.; Cox, J.; Bricker, D.K.; Brockmann, K.; Rutter, J.; Thummel, C.S.; Winge, D.R. The LYR factors SDHAF1 and SDHAF3 mediate maturation of the iron-sulfur subunit of succinate dehydrogenase. Cell Metab. 2014, 20, 253–266. [Google Scholar] [CrossRef] [PubMed]
- Lefebvre-Legendre, L.; Vaillier, J.; Benabdelhak, H.; Velours, J.; Slonimski, P.P.; di Rago, J.P. Identification of a nuclear gene (FMC1) required for the assembly/stability of yeast mitochondrial F(1)-ATPase in heat stress conditions. J. Biol. Chem. 2001, 276, 6789–6796. [Google Scholar] [CrossRef] [PubMed]
- Qiu, J.; Gao, C.L.; Zhang, M.; Chen, R.H.; Chi, X.; Liu, F.; Zhang, C.M.; Ji, C.B.; Chen, X.H.; Zhao, Y.P.; et al. LYRM1, a novel gene promotes proliferation and inhibits apoptosis of preadipocytes. Eur. J. Endocrinol. 2009, 160, 177–184. [Google Scholar]
- Shi, Y.; Ghosh, M.C.; Tong, W.H.; Rouault, T.A. Human ISD11 is essential for both iron-sulfur cluster assembly and maintenance of normal cellular iron homeostasis. Hum. Mol. Genet. 2009, 18, 3014–3025. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.Z.; Zhang, M.; Kou, C.Z.; Ni, Y.H.; Ji, C.B.; Cao, X.G.; Guo, X.R. Effects of LYRM1 knockdown on mitochondrial function in 3 T3-L1 murine adipocytes. J. Bioenerg. Biomembr. 2012, 44, 225–232. [Google Scholar] [CrossRef] [PubMed]
- Haack, T.B.; Madignier, F.; Herzer, M.; Lamantea, E.; Danhauser, K.; Invernizzi, F.; Koch, J.; Freitag, M.; Drost, R.; Hillier, I.; et al. Mutation screening of 75 candidate genes in 152 complex I deficiency cases identifies pathogenic variants in 16 genes including NDUFB9. J. Med. Genet. 2012, 49, 83–89. [Google Scholar]
- Lim, S.C.; Friemel, M.; Marum, J.E.; Tucker, E.J.; Bruno, D.L.; Riley, L.G.; Christodoulou, J.; Kirk, E.P.; Boneh, A.; DeGennaro, C.M.; et al. Mutations in LYRM4, encoding iron-sulfur cluster biogenesis factor ISD11, cause deficiency of multiple respiratory chain complexes. Hum. Mol. Genet. 2013, 22, 4460–4473. [Google Scholar]
- Ladha, J.S.; Tripathy, M.K.; Mitra, D. Mitochondrial complex I activity is impaired during HIV-1-induced T-cell apoptosis. Cell Death. Differ. 2005, 12, 1417–1428. [Google Scholar] [CrossRef] [PubMed]
- Invernizzi, F.; Tigano, M.; Dallabona, C.; Donnini, C.; Ferrero, I.; Cremonte, M.; Ghezzi, D.; Lamperti, C.; Zeviani, M. A homozygous mutation in LYRM7/MZM1L associated with early onset encephalopathy, lactic acidosis, and severe reduction of mitochondrial complex III activity. Hum. Mutat. 2013, 34, 1619–1622. [Google Scholar] [CrossRef] [PubMed]
- Ghezzi, D.; Goffrini, P.; Uziel, G.; Horvath, R.; Klopstock, T.; Lochmuller, H.; D’Adamo, P.; Gasparini, P.; Strom, T.M.; Prokisch, H.; et al. SDHAF1, encoding a LYR complex-II specific assembly factor, is mutated in SDH-defective infantile leukoencephalopathy. Nat. Genet. 2009, 41, 654–656. [Google Scholar]
- Dick, D.M.; Aliev, F.; Wang, J.C.; Saccone, S.; Hinrichs, A.; Bertelsen, S.; Budde, J.; Saccone, N.; Foroud, T.; Nurnberger, J., Jr.; et al. A Systematic single nucleotide polymorphism screen to fine-map alcohol dependence genes on chromosome 7 identifies association with a novel susceptibility gene ACN9. Biol. Psychiatry 2008, 63, 1047–1053. [Google Scholar]
- Richards, T.A.; van der Giezen, M. Evolution of the Isd11-IscS complex reveals a single alpha-proteobacterial endosymbiosis for all eukaryotes. Mol. Biol. Evol. 2006, 23, 1341–1344. [Google Scholar] [CrossRef] [PubMed]
- Stehling, O.; Wilbrecht, C.; Lill, R. Mitochondrial iron-sulfur protein biogenesis and human disease. Biochimie 2014, 100, 61–77. [Google Scholar] [CrossRef] [PubMed]
- Maio, N.; Singh, A.; Uhrigshardt, H.; Saxena, N.; Tong, W.H.; Rouault, T.A. Cochaperone binding to LYR motifs confers specificity of iron sulfur cluster delivery. Cell Metab. 2014, 19, 445–457. [Google Scholar] [CrossRef] [PubMed]
- Cai, K.; Frederick, R.O.; Kim, J.H.; Reinen, N.M.; Tonelli, M.; Markley, J.L. Human mitochondrial chaperone (mtHSP70) and cysteine desulfurase (NFS1) bind preferentially to the disordered conformation, whereas co-chaperone (HSC20) binds to the structured conformation of the iron-sulfur cluster scaffold protein (ISCU). J. Biol. Chem. 2013, 288, 28755–28770. [Google Scholar] [CrossRef] [PubMed]
- Maio, N.; Rouault, T.A. Iron-sulfur cluster biogenesis in mammalian cells: New insights into the molecular mechanisms of cluster delivery. Biochim. Biophys. Acta 2014. [Google Scholar] [CrossRef]
- Claros, M.G.; Vincens, P. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur. J. Biochem. 1996, 241, 779–786. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar]
- Zickermann, V.; Wirth, C.; Nasiri, H.; Siegmund, K.; Schwalbe, H.; Hunte, C.; and Brandt, U. Structural biology. Mechanistic insight from the crystal structure of mitochondrial complex I. Science 2015, 347, 44–49. [Google Scholar]
- Vinothkumar, K.R.; Zhu, J.; Hirst, J. Architecture of mammalian respiratory complex I. Nature 2014, 515, 80–84. [Google Scholar] [CrossRef] [PubMed]
- Angerer, H.; Radermacher, M.; Mankowska, M.; Steger, M.; Zwicker, K.; Heide, H.; Wittig, I.; Brandt, U.; Zickermann, V. The LYR protein subunit NB4M/NDUFA6 of mitochondrial complex I anchors an acyl carrier protein and is essential for catalytic activity. Proc. Natl. Acad. Sci. USA 2014, 111, 5207–5212. [Google Scholar] [CrossRef] [PubMed]
- Hunte, C.; Zickermann, V.; Brandt, U. Functional modules and structural basis of conformational coupling in mitochondrial complex I. Science 2010, 329, 448–451. [Google Scholar] [CrossRef] [PubMed]
- Hirst, J. Mitochondrial complex I. Annu. Rev. Biochem. 2013, 82, 551–575. [Google Scholar] [CrossRef] [PubMed]
- Runswick, M.J.; Fearnley, I.M.; Skehel, J.M.; Walker, J.E. Presence of an acyl carrier protein in NADH:Ubiquinone oxidoreductase from bovine heart mitochondria. FEBS Lett. 1991, 286, 121–124. [Google Scholar] [CrossRef] [PubMed]
- Cronan, J.E.; Fearnley, I.M.; Walker, J.E. Mammalian mitochondria contain a soluble acyl carrier protein. FEBS Lett. 2005, 579, 4892–4896. [Google Scholar] [CrossRef] [PubMed]
- Angerer, H.; Zwicker, K.; Wumaier, Z.; Sokolova, L.; Heide, H.; Steger, M.; Kaiser, S.; Nubel, E.; Brutschy, B.; Radermacher, M.; et al. A scaffold of accessory subunits links the peripheral arm and the distal proton-pumping module of mitochondrial complex I. Biochem. J. 2011, 437, 279–288. [Google Scholar]
- Dobrynin, K.; Abdrakhmanova, A.; Richers, S.; Hunte, C.; Kerscher, S.; Brandt, U. Characterization of two different acyl carrier proteins in complex I from Yarrowia lipolytica. Biochim. Biophys. Acta 2010, 1797, 152–159. [Google Scholar] [CrossRef] [PubMed]
- Feng, D.; Witkowski, A.; Smith, S. Down-regulation of mitochondrial acyl carrier protein in mammalian cells compromises protein lipoylation and respiratory complex I and results in cell death. J. Biol. Chem. 2009, 284, 11436–11445. [Google Scholar] [CrossRef] [PubMed]
- Hiltunen, J.K.; Autio, K.J.; Schonauer, M.S.; Kursu, V.A.; Dieckmann, C.L.; Kastaniotis, A.J. Mitochondrial fatty acid synthesis and respiration. Biochim. Biophys. Acta 2010, 1797, 1195–1202. [Google Scholar] [CrossRef] [PubMed]
- Schneider, R.; Massow, M.; Lisowsky, T.; Weiss, H. Different respiratory-defective phenotypes of Neurospora crassa and Saccharomyces cerevisiae after inactivation of the gene encoding the mitochondrial acyl carrier protein. Curr. Genet. 1995, 29, 10–17. [Google Scholar] [CrossRef] [PubMed]
- Goraca, A.; Huk-Kolega, H.; Piechota, A.; Kleniewska, P.; Ciejka, E.; Skibska, B. Lipoic acid-biological activity and therapeutic potential. Pharmacol. Rep. 2011, 63, 849–858. [Google Scholar] [CrossRef] [PubMed]
- Yi, X.; Maeda, N. Endogenous production of lipoic acid is essential for mouse development. Mol. Cell Biol. 2005, 25, 8387–8392. [Google Scholar] [CrossRef] [PubMed]
- Carroll, J.; Fearnley, I.M.; Shannon, R.J.; Hirst, J.; Walker, J.E. Analysis of the subunit composition of complex I from bovine heart mitochondria. Mol. Cell Proteomics. 2003, 2, 117–126. [Google Scholar] [CrossRef] [PubMed]
- Kmita, K.; Zickermann, V. Accessory subunits of mitochondrial complex I. Biochem. Soc. Trans. 2013, 41, 1272–1279. [Google Scholar] [CrossRef] [PubMed]
- Babot, M.; Birch, A.; Labarbuta, P.; Galkin, A. Characterisation of the active/de-active transition of mitochondrial complex I. Biochim. Biophys. Acta 2014, 1837, 1083–1092. [Google Scholar] [CrossRef] [PubMed]
- Brody, S.; Mikolajczyk, S. Neurospora mitochondria contain an acyl-carrier protein. Eur. J. Biochem. 1988, 173, 353–359. [Google Scholar] [CrossRef] [PubMed]
- Franceschini, A.; Szklarczyk, D.; Frankild, S.; Kuhn, M.; Simonovic, M.; Roth, A.; Lin, J.; Minguez, P.; Bork, P.; von, M.C.; et al. STRING v9.1: Protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013, 41, D808–D815. [Google Scholar]
- Krogan, N.J.; Cagney, G.; Yu, H.; Zhong, G.; Guo, X.; Ignatchenko, A.; Li, J.; Pu, S.; Datta, N.; Tikuisis, A.P.; et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 2006, 440, 637–643. [Google Scholar]
- Guruharsha, K.G.; Rual, J.F.; Zhai, B.; Mintseris, J.; Vaidya, P.; Vaidya, N.; Beekman, C.; Wong, C.; Rhee, D.Y.; Cenaj, O.; et al. A protein complex network of Drosophila melanogaster. Cell 2011, 147, 690–703. [Google Scholar] [CrossRef]
- Lill, R.; Hoffmann, B.; Molik, S.; Pierik, A.J.; Rietzschel, N.; Stehling, O.; Uzarska, M.A.; Webert, H.; Wilbrecht, C.; Muhlenhoff, U. The role of mitochondria in cellular iron-sulfur protein biogenesis and iron metabolism. Biochim. Biophys. Acta 2012, 1823, 1491–1508. [Google Scholar] [CrossRef] [PubMed]
- Shan, Y.; Napoli, E.; Cortopassi, G. Mitochondrial frataxin interacts with ISD11 of the NFS1/ISCU complex and multiple mitochondrial chaperones. Hum. Mol. Genet. 2007, 16, 929–941. [Google Scholar] [CrossRef] [PubMed]
- Wiedemann, N.; Urzica, E.; Guiard, B.; Muller, H.; Lohaus, C.; Meyer, H.E.; Ryan, M.T.; Meisinger, C.; Muhlenhoff, U.; Lill, R.; et al. Essential role of Isd11 in mitochondrial iron-sulfur cluster synthesis on Isu scaffold proteins. EMBO J. 2006, 25, 184–195. [Google Scholar]
- Pandey, A.; Golla, R.; Yoon, H.; Dancis, A.; Pain, D. Persulfide formation on mitochondrial cysteine desulfurase: Enzyme activation by a eukaryote-specific interacting protein and Fe-S cluster synthesis. Biochem. J. 2012, 448, 171–187. [Google Scholar] [CrossRef] [PubMed]
- Terali, K.; Beavil, R.L.; Pickersgill, R.W.; van der Giezen, M. The effect of the adaptor protein Isd11 on the quaternary structure of the eukaryotic cysteine desulphurase Nfs1. Biochem. Biophys. Res. Commun. 2013, 440, 235–240. [Google Scholar] [CrossRef] [PubMed]
- Lu, K.Y.; Tao, S.C.; Yang, T.C.; Ho, Y.H.; Lee, C.H.; Lin, C.C.; Juan, H.F.; Huang, H.C.; Yang, C.Y.; Chen, M.S.; et al. Profiling lipid-protein interactions using nonquenched fluorescent liposomal nanovesicles and proteome microarrays. Mol. Cell Proteomics. 2012, 11, 1177–1190. [Google Scholar]
- Cicchillo, R.M.; Booker, S.J. Mechanistic investigations of lipoic acid biosynthesis in Escherichia coli: Both sulfur atoms in lipoic acid are contributed by the same lipoyl synthase polypeptide. J. Am. Chem. Soc. 2005, 127, 2860–2861. [Google Scholar] [CrossRef] [PubMed]
- Harmer, J.E.; Hiscox, M.J.; Dinis, P.C.; Fox, S.J.; Iliopoulos, A.; Hussey, J.E.; Sandy, J.; van Beek, F.T.; Essex, J.W.; Roach, P.L. Structures of lipoyl synthase reveal a compact active site for controlling sequential sulfur insertion reactions. Biochem. J. 2014, 464, 123–133. [Google Scholar] [CrossRef] [PubMed]
- Gerber, J.; Muhlenhoff, U.; Lill, R. An interaction between frataxin and Isu1/NFS1 that is crucial for Fe/S cluster synthesis on Isu1. EMBO Rep. 2003, 4, 906–911. [Google Scholar] [CrossRef] [PubMed]
- Condo, I.; Malisan, F.; Guccini, I.; Serio, D.; Rufini, A.; Testi, R. Molecular control of the cytosolic aconitase/IRP1 switch by extramitochondrial frataxin. Hum. Mol. Genet. 2010, 19, 1221–1229. [Google Scholar] [CrossRef] [PubMed]
- Bulteau, A.L.; O’Neill, H.A.; Kennedy, M.C.; Ikeda-Saito, M.; Isaya, G.; Szweda, L.I. Frataxin acts as an iron chaperone protein to modulate mitochondrial aconitase activity. Science 2004, 305, 242–245. [Google Scholar] [CrossRef] [PubMed]
- Mimaki, M.; Wang, X.; McKenzie, M.; Thorburn, D.R.; Ryan, M.T. Understanding mitochondrial complex I assembly in health and disease. Biochim. Biophys. Acta 2012, 1817, 851–862. [Google Scholar] [CrossRef] [PubMed]
- Newgard, C.B. Interplay between lipids and branched-chain amino acids in development of insulin resistance. Cell Metab. 2012, 15, 606–614. [Google Scholar] [CrossRef] [PubMed]
- Lynch, C.J.; Adams, S.H. Branched-chain amino acids in metabolic signalling and insulin resistance. Nat. Rev. Endocrinol. 2014, 10, 723–736. [Google Scholar] [CrossRef] [PubMed]
- Kniazeva, M.; Euler, T.; Han, M. A branched-chain fatty acid is involved in post-embryonic growth control in parallel to the insulin receptor pathway and its biosynthesis is feedback-regulated in C. elegans. Genes Dev. 2008, 22, 2102–2110. [Google Scholar] [CrossRef]
- Kniazeva, M.; Shen, H.; Euler, T.; Wang, C.; Han, M. Regulation of maternal phospholipid composition and IP(3)-dependent embryonic membrane dynamics by a specific fatty acid metabolic event in C. elegans. Genes Dev. 2012, 26, 554–566. [Google Scholar] [CrossRef]
- Lin, T.; Yin, X.; Cai, Q.; Fan, X.; Xu, K.; Huang, L.; Luo, J.; Zheng, J.; Huang, J. 13-Methyltetradecanoic acid induces mitochondrial-mediated apoptosis in human bladder cancer cells. Urol. Oncol. Semin. Orig. Investig. 2012, 30, 339–345. [Google Scholar] [CrossRef]
- Zensen, R.; Husmann, H.; Schneider, R.; Peine, T.; Weiss, H. De novo synthesis and desaturation of fatty acids at the mitochondrial acyl-carrier protein, a subunit of NADH:Ubiquinone oxidoreductase in Neurospora crassa. FEBS Lett. 1992, 310, 179–181. [Google Scholar] [CrossRef] [PubMed]
- Lancaster, C.R.; Kroger, A. Succinate: Quinone oxidoreductases: New insights from X-ray crystal structures. Biochim. Biophys. Acta 2000, 1459, 422–431. [Google Scholar] [CrossRef] [PubMed]
- Drose, S.; Brandt, U. Molecular mechanisms of superoxide production by the mitochondrial respiratory chain. Adv. Exp. Med. Biol. 2012, 748, 145–169. [Google Scholar] [PubMed]
- Crofts, A.R. The cytochrome bc1 complex: Function in the context of structure. Annu. Rev. Physiol. 2004, 66, 689–733. [Google Scholar] [CrossRef] [PubMed]
- Atkinson, A.; Smith, P.; Fox, J.L.; Cui, T.Z.; Khalimonchuk, O.; Winge, D.R. The LYR protein Mzm1 functions in the insertion of the Rieske Fe/S protein in yeast mitochondria. Mol. Cell Biol. 2011, 31, 3988–3996. [Google Scholar] [CrossRef] [PubMed]
- Cui, T.Z.; Smith, P.M.; Fox, J.L.; Khalimonchuk, O.; Winge, D.R. Late-stage maturation of the Rieske Fe/S protein: Mzm1 stabilizes Rip1 but does not facilitate its translocation by the AAA ATPase Bcs1. Mol. Cell Biol. 2012, 32, 4400–4409. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, E.; Lobo, T.; Fox, J.L.; Zeviani, M.; Winge, D.R.; Fernandez-Vizarra, E. LYRM7/MZM1L is a UQCRFS1 chaperone involved in the last steps of mitochondrial complex III assembly in human cells. Biochim. Biophys. Acta 2013, 1827, 285–293. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Cheng, G.; Zhu, H.; Guan, G. A study of genes involved in adipocyte differentiation. J. Pediatr. Endocrinol. Metab 2014. [Google Scholar]
- Yin, C.; Xiao, Y.; Zhang, W.; Xu, E.; Liu, W.; Yi, X.; Chang, M. DNA microarray analysis of genes differentially expressed in adipocyte differentiation. J. Biosci. 2014, 39, 415–423. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.G.; Kou, C.Z.; Zhao, Y.P.; Gao, C.L.; Zhu, C.; Zhang, C.M.; Ji, C.B.; Qin, D.N.; Zhang, M.; Guo, X.R. Overexpression of LYRM1 induces mitochondrial impairment in 3T3-L1 adipocytes. Mol. Genet. Metab. 2010, 101, 395–399. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Qin, Z.Y.; Dai, Y.M.; Wang, Y.M.; Zhu, G.Z.; Zhao, Y.P.; Ji, C.B.; Zhu, J.G.; Shi, C.M.; Qiu, J.; et al. Knockdown of LYRM1 rescues insulin resistance and mitochondrial dysfunction induced by FCCP in 3T3-L1 adipocytes. Cell Biochem. Biophys. 2014, 70, 667–675. [Google Scholar]
- Kou, C.; Cao, X.; Qin, D.; Ji, C.; Zhu, J.; Zhang, C.; Zhu, C.; Gao, C.; Chen, R.; Guo, X.; et al. Over-expression of LYRM1 inhibits glucose transport in rat skeletal muscles via attenuated phosphorylation of PI3K (p85) and Akt. Mol. Cell Biochem. 2011, 348, 149–154. [Google Scholar]
- Qin, Z.Y.; Zhang, M.; Guo, X.R.; Wang, Y.M.; Zhu, G.Z.; Ni, Y.H.; Zhao, Y.P.; Qiu, J.; Kou, C.Z.; Qin, R.; et al. Alpha-lipoic acid ameliorates impaired glucose uptake in LYRM1 overexpressing 3T3-L1 adipocytes through the IRS-1/Akt signaling pathway. J. Bioenerg. Biomembr. 2012, 44, 579–586. [Google Scholar]
- Qin, Z.Y.; Zhang, M.; Dai, Y.M.; Wang, Y.M.; Zhu, G.Z.; Zhao, Y.P.; Ji, C.B.; Qiu, J.; Cao, X.G.; Guo, X.R. Metformin prevents LYRM1-induced insulin resistance in 3T3-L1 adipocytes via a mitochondrial-dependent mechanism. Exp. Biol. Med. (Maywood.) 2014, 239, 1567–1574. [Google Scholar] [CrossRef]
- Bridges, H.R.; Jones, A.J.; Pollak, M.N.; Hirst, J. Effects of metformin and other biguanides on oxidative phosphorylation in mitochondria. Biochem. J. 2014, 462, 475–487. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Shao, J.; Li, H.; Yu, Y. Temporal gene expression changes induced by a low concentration of benzo[a]pyrene diol epoxide in a normal human cell line. Mutat. Res. 2010, 684, 74–80. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.Y.; Fridley, B.L.; Jenkins, G.D.; Batzler, A.; Wang, L.; Weinshilboum, R.M. Mycophenolic acid response biomarkers: A cell line model system-based genome-wide screen. Int. Immunopharmacol. 2011, 11, 1057–1064. [Google Scholar] [CrossRef] [PubMed]
- Van der Valk, R.J.; Duijts, L.; Timpson, N.J.; Salam, M.T.; Standl, M.; Curtin, J.A.; Genuneit, J.; Kerhof, M.; Kreiner-Moller, E.; Caceres, A.; et al. Fraction of exhaled nitric oxide values in childhood are associated with 17q11.2-q12 and 17q12-q21 variants. J. Allergy Clin. Immunol. 2014, 134, 46–55. [Google Scholar]
- Khalimonchuk, O.; Ott, M.; Funes, S.; Ostermann, K.; Rodel, G.; Herrmann, J.M. Sequential processing of a mitochondrial tandem protein: Insights into protein import in Schizosaccharomyces pombe. Eukaryot. Cell 2006, 5, 997–1006. [Google Scholar] [CrossRef] [PubMed]
- Chakrabarti, V.S.; Mikolajczyk, M.; Boscaro, F.; Calderone, V. Human Ind1 expression causes over-expression of E. coli beta-lactamase ampicillin resistance protein. Protein Expr. Purif. 2014, 104C, 26–33. [Google Scholar]
- Wydro, M.M.; Sharma, P.; Foster, J.M.; Bych, K.; Meyer, E.H.; Balk, J. The evolutionarily conserved iron-sulfur protein INDH is required for complex I assembly and mitochondrial translation in Arabidopsis (corrected). Plant Cell 2013, 25, 4014–4027. [Google Scholar] [CrossRef] [PubMed]
- Calvo, S.E.; Tucker, E.J.; Compton, A.G.; Kirby, D.M.; Crawford, G.; Burtt, N.P.; Rivas, M.; Guiducci, C.; Bruno, D.L.; Goldberger, O.A.; et al. High-throughput, pooled sequencing identifies mutations in NUBPL and FOXRED1 in human complex I deficiency. Nat. Genet. 2010, 42, 851–858. [Google Scholar]
- Boratyn, G.M.; Camacho, C.; Cooper, P.S.; Coulouris, G.; Fong, A.; Ma, N.; Madden, T.L.; Matten, W.T.; McGinnis, S.D.; Merezhuk, Y.; et al. BLAST: A more efficient report with usability improvements. Nucleic Acids Res. 2013, 41, W29–W33. [Google Scholar]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Angerer, H. Eukaryotic LYR Proteins Interact with Mitochondrial Protein Complexes. Biology 2015, 4, 133-150. https://doi.org/10.3390/biology4010133
Angerer H. Eukaryotic LYR Proteins Interact with Mitochondrial Protein Complexes. Biology. 2015; 4(1):133-150. https://doi.org/10.3390/biology4010133
Chicago/Turabian StyleAngerer, Heike. 2015. "Eukaryotic LYR Proteins Interact with Mitochondrial Protein Complexes" Biology 4, no. 1: 133-150. https://doi.org/10.3390/biology4010133
APA StyleAngerer, H. (2015). Eukaryotic LYR Proteins Interact with Mitochondrial Protein Complexes. Biology, 4(1), 133-150. https://doi.org/10.3390/biology4010133