Color Mutations Alter the Biochemical Composition in the San Marzano Tomato Fruit
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material and Growth Conditions
2.2. Fruit Sampling
2.3. Volatile Detection and Quantification
2.4. Non-Volatile Detection and Quantification
2.5. Statistical and Bioinformatics Analyses
3. Results
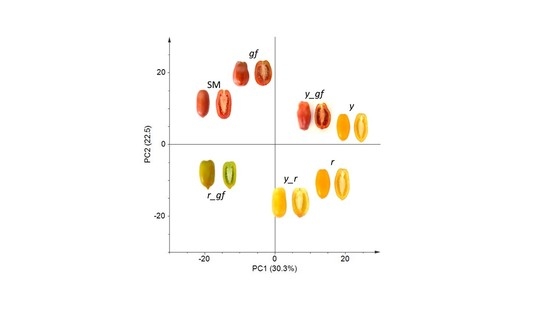
3.1. Untargeted Analysis of Volatile, Non-Polar and Polar Metabolites
3.2. Estimation of “Gen*Year” Interaction in the Quantification of Targeted Metabolites
3.3. Targeted Analysis of Volatile Compounds
3.4. Targeted Analysis of Non-Polar Metabolites
3.5. Targeted Analysis of Polar Metabolites
3.6. Bioinformatics to Investigate Metabolite-Metabolite Relationships
4. Discussion
4.1. Biochemical Changes in Fruits of Yellow Flesh and of Its Combinations with y and gf
4.2. Biochemical Changes in Fruits of Colorless Fruit Epidermis and of Its Combinations with r and gf
4.3. Biochemical Changes in Fruits of Green Flesh and of Its Combinations with r and y
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lewinsohn, E.; Sitrit, Y.; Bar, E.; Azulay, Y.; Ibdah, M.; Meir, A.; Yosef, E.; Zamir, D.; Tadmor, Y. Not just colors—carotenoid degradation as a link between pigmentation and aroma in tomato and watermelon fruit. Trends Food Sci. Technol. 2005, 16, 407–415. [Google Scholar] [CrossRef]
- Gascuel, Q.; Diretto, G.; Monforte, A.J.; Fortes, A.M.; Granell, A. Use of natural diversity and biotechnology to increase the quality and nutritional content of tomato and grape. Front. Plant Sci. 2017, 8, 652. [Google Scholar] [CrossRef] [Green Version]
- Bovy, A.G.; Gómez-Roldán, V.; Hall, R.D. Strategies to optimize the flavonoid content of tomato fruit. Recent Adv. Polyphen. Res. 2010, 2, 138–162. [Google Scholar]
- Hunt, G.M.; Baker, E.A. Phenolic constituents of tomato fruit cuticles. Phytochemistry 1980, 19, 1415–1419. [Google Scholar] [CrossRef]
- Dixon, R.A. Natural products and plant disease resistance. Nature 2001, 411, 843–847. [Google Scholar] [CrossRef]
- Sands, D.C.; Morris, C.E.; Dratz, E.A.; Pilgeram, A. Elevating optimal human nutrition to a central goal of plant breeding and production of plant-based foods. Plant Sci. 2009, 177, 377–389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, C.; Li, J. Medicine is not health care, food is health care: Plant metabolic engineering, diet and human health. New Phytol. 2017, 216, 699–719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martínez-Valverde, I.; Periago, M.J.; Provan, G.; Chesson, A. Phenolic compounds, lycopene and antioxidant activity in commercial varieties of tomato (Lycopersicum esculentum). J. Sci. Food Agric. 2002, 82, 323–330. [Google Scholar] [CrossRef]
- Jenkins, J.A.; Mackinney, G. Carotenoids of the apricot tomato and its hybrids with yellow and tangerine. Genetics 1955, 40, 715. [Google Scholar]
- Rodríguez-Villalón, A.; Gas, E.; Rodríguez-Concepción, M. Phytoene synthase activity controls the biosynthesis of carotenoids and the supply of their metabolic precursors in dark-grown Arabidopsis seedlings. Plant J. 2009, 60, 424–435. [Google Scholar] [CrossRef]
- Kachanovsky, D.E.; Filler, S.; Isaacson, T.; Hirschberg, J. Epistasis in tomato color mutations involves regulation of phytoene synthase 1 expression by cis-carotenoids. Proc. Natl. Acad. Sci. USA 2012, 109, 19021–19026. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rocha, M.D.C.; Deliza, R.; Corrêa, F.M.; do Carmo, M.G.; Abboud, A.C. A study to guide breeding of new cultivars of organic cherry tomato following a consumer-driven approach. Food Res. 2013, 51, 265–273. [Google Scholar] [CrossRef] [Green Version]
- Lindstrom, E.W. Inheritance in tomato. Genetica 1925, 10, 305–317. [Google Scholar]
- Rick, C.M.; Butler, L. Cytogenetics of the tomato. Adv. Genet. 1956, 8, 267–382. [Google Scholar]
- Adato, A.; Mandel, T.; Mintz-Oron, S.; Venger, I.; Levy, D.; Yativ, M.; Domìnguez, E.; Wang, Z.; De Vos, R.C.; Jetter, R.; et al. Fruit-surface flavonoid accumulation in tomato is controlled by a SlMYB12-regulated transcriptional network. PLoS Genet. 2009, 5, e1000777. [Google Scholar] [CrossRef] [Green Version]
- Ballester, A.R.; Molthoff, J.; de Vos, R.; Hekkert, B.T.; Orzaez, D.; Fernández-Moreno, J.P.; Tripodi, P.; Grandillo, S.; Martin, C.; Heldens, J.; et al. Biochemical and molecular analysis of pink tomatoes: Deregulated expression of the gene encoding transcription factor SlMYB12 leads to pink tomato fruit color. Plant Physiol. 2010, 152, 71–84. [Google Scholar] [CrossRef] [Green Version]
- Kerr, E.A. Green flesh, gf. Rep. Tomato Genet. Coop. 1956, 6, 17. [Google Scholar]
- Barry, C.S.; McQuinn, R.P.; Chung, M.Y.; Besuden, A.; Giovannoni, J.J. Amino acid substitutions in homologs of the STAY-GREEN protein are responsible for the green-flesh and chlorophyll retainer mutations of tomato and pepper. Plant Physiol. 2008, 147, 179–187. [Google Scholar] [CrossRef] [Green Version]
- Barry, C.S.; Cornelius, S.; Pandey, P. A survey of cultivated heirloom tomato varieties identifies four new mutant alleles at the green-flesh locus. Mol. Breed. 2009, 24, 269–276. [Google Scholar] [CrossRef]
- Bortolotti, S.; Boggio, S.B.; Delgado, L.; Orellano, E.G.; Valle, E.M. Different induction patterns of glutamate metabolizing enzymes in ripening fruits of the tomato mutant green flesh. Physiol. Plant. 2003, 119, 384–391. [Google Scholar] [CrossRef]
- Cocaliadis, M.F.; Fernández-Muñoz, R.; Pons, C.; Orzaez, D.; Granell, A. Increasing tomato fruit quality by enhancing fruit chloroplast function. A double-edged sword? J. Exp. Bot. 2014, 65, 4589–4598. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loiudice, R.; Impembo, M.; Laratta, B.; Villari, G.; Voi, A.L.; Siviero, P.; Castaldo, D. Composition of San Marzano tomato varieties. Food Chem. 1995, 53, 81–89. [Google Scholar] [CrossRef]
- Ercolano, M.R.; Carli, P.; Soria, A.; Cascone, A.; Fogliano, V.; Frusciante, L.; Barone, A. Biochemical, sensorial and genomic profiling of traditional Italian tomato varieties. Euphytica 2008, 164, 571–582. [Google Scholar] [CrossRef]
- D’Esposito, D.; Ferriello, F.; Dal Molin, A.; Diretto, G.; Sacco, A.; Minio, A.; Barone, A.; Di Monaco, R.; Cavella, S.; Tardella, L.; et al. Unravelling the complexity of transcriptomic, metabolomic and quality environmental response of tomato fruit. BMC Plant Biol. 2017, 17, 66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barone, D.; Cito, L.; Tommonaro, G.; Abate, A.A.; Penon, D.; De Prisco, R.; Penon, A.; Forte, I.M.; Benedetti, E.; Cimini, A.; et al. Antitumoral potential, antioxidant activity and carotenoid content of two Southern Italy tomato cultivars extracts: San Marzano and Corbarino. J. Cell Physiol. 2018, 233, 1266–1277. [Google Scholar] [CrossRef]
- Dono, G.; Picarella, M.E.; Pons, C.; Santangelo, E.; Monforte, A.; Granell, A.; Mazzucato, A. Characterization of a repertoire of tomato fruit genetic variants in the San marzano genetic background. Sci. Hortic. 2020, 261, 108927. [Google Scholar] [CrossRef]
- Rambla, J.L.; Medina, A.; Fernández-del-Carmen, A.; Barrantes, W.; Grandillo, S.; Cammareri, M.; Lopez-Casado, G.; Rodrigo, G.; Alonso, A.; Garcìa-Martinez, S. Identification, introgression, and validation of fruit volatile QTLs from a red-fruited wild tomato species. J. Exp. Bot. 2017, 68, 429–442. [Google Scholar] [CrossRef]
- Tikunov, Y.; Lommen, A.; De Vos, C.R.; Verhoeven, H.A.; Bino, R.J.; Hall, R.D.; Bovy, A.G. A novel approach for nontargeted data analysis for metabolomics. Large-scale profiling of tomato fruit volatiles. Plant Physiol. 2005, 139, 1125–1137. [Google Scholar] [CrossRef] [Green Version]
- Fasano, C.; Diretto, G.; Aversano, R.; D’Agostino, N.; Di Matteo, A.; Frusciante, L.; Giuliano, G.; Carputo, D. Transcriptome and metabolome of synthetic Solanum autotetraploids reveal key genomic stress events following polyploidization. New Phytol. 2016, 210, 1382–1394. [Google Scholar] [CrossRef] [Green Version]
- Rambla, J.L.; Trapero-Mozos, A.; Diretto, G.; Rubio-Moraga, A.; Granell, A.; Gómez-Gómez, L.; Ahrazem, O. Gene-metabolite networks of volatile metabolism in Airen and Tempranillo grape cultivars revealed a distinct mechanism of aroma bouquet production. Front. Plant Sci. 2016, 7, 1619. [Google Scholar] [CrossRef] [Green Version]
- Diretto, G.; Rubio-Moraga, A.; Argandoña, J.; Castillo, P.; Gómez-Gómez, L.; Ahrazem, O. Tissue-specific accumulation of sulfur compounds and saponins in different parts of garlic cloves from purple and white ecotypes. Molecules 2017, 22, 1359. [Google Scholar] [CrossRef] [PubMed]
- Sulli, M.; Mandolino, G.; Sturaro, M.; Onofri, C.; Diretto, G.; Parisi, B.; Giuliano, G. Molecular and biochemical characterization of a potato collection with contrasting tuber carotenoid content. PLoS One 2017, 12, e0184143. [Google Scholar] [CrossRef] [PubMed]
- Cappelli, G.; Giovannini, D.; Basso, A.L.; Demurtas, O.C.; Diretto, G.; Santi, C.; Girelli, G.; Bacchetta, L.; Mariani, F. A Corylus avellana L. extract enhances human macrophage bactericidal response against Staphylococcus aureus by increasing the expression of anti-inflammatory and iron metabolism genes. J. Funct. Foods 2018, 45, 499–511. [Google Scholar] [CrossRef]
- Ahrazem, O.; Argandoña, J.; Fiore, A.; Aguado, C.; Luján, R.; Rubio-Moraga, Á.; Marro, M.; Araujo-Andrade, C.; Loza-Alvarez, P.; Diretto, G.; et al. Transcriptome analysis in tissue sectors with contrasting crocins accumulation provides novel insights into apocarotenoid biosynthesis and regulation during chromoplast biogenesis. Sci. Rep. 2018, 8, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Diretto, G.; Frusciante, S.; Fabbri, C.; Schauer, N.; Busta, L.; Wang, Z.; Matas, A.J.; Fiore, A.; Rose, J.K.C.; Fernie, R.A. Manipulation of β-carotene levels in tomato fruits results in increased ABA content and extended shelf life. Plant Biotechnol. J. 2019, in press. [Google Scholar] [CrossRef] [PubMed]
- Bader, G.D.; Hogue, C.W. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinf. 2003, 4, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tieman, D.; Zhu, G.; Resende, M.F.; Lin, T.; Nguyen, C.; Bies, D.; Rambla, J.L.; Stephanie Ortiz Beltran, K.; Taylor, M. A chemical genetic roadmap to improved tomato flavor. Science 2017, 355, 391–394. [Google Scholar] [CrossRef]
- Rodríguez-Burruezo, A.; Prohens, J.; Roselló, S.; Nuez, F. “Heirloom” varieties as sources of variation for the improvement of fruit quality in greenhouse-grown tomatoes. J Hort. Sci. Biotechnol. 2005, 80, 453–460. [Google Scholar] [CrossRef]
- Flores, P.; Sánchez, E.; Fenoll, J.; Hellín, P. Genotypic variability of carotenoids in traditional tomato cultivars. Food Res. Int. 2017, 100, 510–516. [Google Scholar] [CrossRef]
- Kang, S.I.; Hwang, I.; Goswami, G.; Jung, H.J.; Nath, U.K.; Yoo, H.J.; Lee, J.M.; Nou, I.S. Molecular insights reveal Psy1, SGR, and SlMYB12 genes are associated with diverse fruit color pigments in tomato (Solanum lycopersicum L.). Molecules 2017, 22, 2180. [Google Scholar] [CrossRef] [Green Version]
- Dal Santo, S.; Fasoli, M.; Negri, S.; D’Incà, E.; Vicenzi, N.; Guzzo, F. Plasticity of the berry ripening program in a white grape variety. Front. Plant Sci. 2016, 7, 970. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- LeRosen, A.L.; Went, F.W.; Zechmeister, L. Relation between genes and carotenoids of the tomato. Proc. Natl. Acad. Sci. USA 1941, 27, 236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, B.; Gu, Q.; Tian, P.; Xiao, L.; Cao, H.; Yang, W. A chimeric transcript containing Psy1 and a potential mRNA is associated with yellow flesh color in tomato accession PI 114490. Planta 2014, 240, 1011–1021. [Google Scholar] [CrossRef] [PubMed]
- Gady, A.L.; Vriezen, W.H.; Van de Wal, M.H.; Huang, P.; Bovy, A.G.; Visser, R.G.; Bachem, C.W. Induced point mutations in the phytoene synthase 1 gene cause differences in carotenoid content during tomato fruit ripening. Mol. Breed. 2012, 29, 801–812. [Google Scholar] [CrossRef] [Green Version]
- Demmig-Adams, B.; Adams Iii, W.W. Photoprotection and other responses of plants to high light stress. Ann. Rev. Plant Biol. 1992, 4, 599–626. [Google Scholar] [CrossRef]
- Parry, A.D.; Babiano, M.J.; Horgan, R. The role of cis-carotenoids in abscisic acid biosynthesis. Planta 1990, 182, 118–128. [Google Scholar] [CrossRef]
- Nohl, H.; Jordan, W.; Youngman, R.J. Quinones in biology: Functions in electron transfer and oxygen activation. Free Radic. Biol. Med. 1986, 2, 211–279. [Google Scholar] [CrossRef]
- Ghorbanli, M.; Gafarabad, M.; Amirkian, T.; Bahareh, A.M. Investigation of proline, total protein, chlorophyll, ascorbate and dehydroascorbate changes under drought stress in Akria and Mobil tomato cultivars. Iran. J. Plant Physiol. 2013, 3, 651–658. [Google Scholar]
- Pál, M.; Tajti, J.; Szalai, G.; Peeva, V.; Végh, B.; Janda, T. Interaction of polyamines, abscisic acid and proline under osmotic stress in the leaves of wheat plants. Sci. Rep. 2018, 8, 1–12. [Google Scholar] [CrossRef]
- Alferez, F.M.; Gerberich, K.M.; Li, J.L.; Zhang, Y.; Graham, J.H.; Mou, Z. Exogenous nicotinamide adenine dinucleotide induces resistance to citrus canker in citrus. Front. Plant Sci. 2018, 9, 1472. [Google Scholar] [CrossRef]
- Vogel, J.T.; Tieman, D.M.; Sims, C.A.; Odabasi, A.Z.; Clark, D.G.; Klee, H.J. Carotenoid content impacts flavor acceptability in tomato (Solanum lycopersicum). J. Sci. Food Agric. 2010, 90, 2233–2240. [Google Scholar] [CrossRef] [PubMed]
- Fray, R.G.; Grierson, D. Identification and genetic analysis of normal and mutant phytoene synthase genes of tomato by sequencing, complementation and co-suppression. Plant Mol. Biol. 1993, 22, 589–602. [Google Scholar] [CrossRef] [PubMed]
- Vogel, J.T.; Tan, B.C.; McCarty, D.R.; Klee, H.J. The carotenoid cleavage dioxygenase 1 enzyme has broad substrate specificity, cleaving multiple carotenoids at two different bond positions. J. Biol. Chem. 2008, 283, 11364–11373. [Google Scholar] [CrossRef] [Green Version]
- Koeduka, T.; Fridman, E.; Gang, D.R.; Vassão, D.G.; Jackson, B.L.; Kish, C.M.; Orlova, I.; Spassova, S.M.; Lewis, N.G.; Noel, J.P.; et al. Eugenol and isoeugenol, characteristic aromatic constituents of spices, are biosynthesized via reduction of a coniferyl alcohol ester. Proc. Natl. Acad. Sci. USA 2006, 103, 10128–10133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borrelli, G.M.; Troccoli, A.; Di Fonzo, N.; Fares, C. Durum wheat lipoxygenase activity and other quality parameters that affect pasta color. Cereal Chem. 1999, 76, 335–340. [Google Scholar] [CrossRef]
- Biehler, E.; Alkerwi, A.A.; Hoffmann, L.; Krause, E.; Guillaume, M.; Lair, M.L.; Bohn, T. Contribution of violaxanthin, neoxanthin, phytoene and phytofluene to total carotenoid intake: Assessment in Luxembourg. J. Food Compos. Anal. 2012, 25, 56–65. [Google Scholar] [CrossRef]
- Liu, M.; Lu, S. Plastoquinone and ubiquinone in plants: Biosynthesis, physiological function and metabolic engineering. Front. Plant Sci. 2016, 7, 1898. [Google Scholar] [CrossRef] [Green Version]
- Mintz-Oron, S.; Mandel, T.; Rogachev, I.; Feldberg, L.; Lotan, O.; Yativ, M.; Jetter, R.; Venger, I.; Adato, A.; Aharoni, A. Gene expression and metabolism in tomato fruit surface tissues. Plant Physiol. 2008, 147, 823–851. [Google Scholar] [CrossRef] [Green Version]
- Fernandez-Moreno, J.P.; Tzfadia, O.; Forment, J.; Presa, S.; Rogachev, I.; Meir, S.; Orzaez, D.; Aharoni, A.; Granell, A. Characterization of a new pink-fruited tomato mutant results in the identification of a null allele of the SlMYB12 transcription factor. Plant Physiol. 2016, 171, 1821–1836. [Google Scholar] [CrossRef] [Green Version]
- Tieman, D.; Taylor, M.; Schauer, N.; Fernie, A.R.; Hanson, A.D.; Klee, H.J. Tomato aromatic amino acid decarboxylases participate in synthesis of the flavor volatiles 2-phenylethanol and 2-phenylacetaldehyde. Proc. Natl. Acad. Sci. USA 2006, 103, 8287–8292. [Google Scholar] [CrossRef] [Green Version]
- Baldwin, E.A.; Goodner, K.; Plotto, A. Interactions of volatiles, sugars, and acids on perception of tomato aroma and flavor descriptors. J. Food Sci. 2008, 73, S294–S307. [Google Scholar] [CrossRef] [PubMed]
- Rambla, J.L.; Tikunov, Y.M.; Monforte, A.J.; Bovy, A.G.; Granell, A. The expanded tomato fruit volatile landscape. J. Exp. Bot. 2014, 65, 4613–4623. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, G.; Hackett, R.; Walker, D.; Taylor, A.; Lin, Z.; Grierson, D. Identification of a specific isoform of tomato lipoxygenase (TomloxC) involved in the generation of fatty acid-derived flavor compounds. Plant Physiol. 2004, 136, 2641–2651. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tieman, D.; Bliss, P.; McIntyre, L.M.; Blandon-Ubeda, A.; Bies, D.; Odabasi, A.Z.; Rodriguez, G.R.; van der Knaap, E.; Taylor, M.G.; Goulet, C. The chemical interactions underlying tomato flavor preferences. Curr. Biol. 2012, 22, 1035–1039. [Google Scholar] [CrossRef] [Green Version]
- Schneider, C. Chemistry and biology of vitamin E. Mol. Nutr. Food Res. 2005, 49, 7–30. [Google Scholar] [CrossRef]
- Domínguez, E.; España, L.; López-Casado, G.; Cuartero, J.; Heredia, A. Biomechanics of isolated tomato (Solanum lycopersicum) fruit cuticles during ripening: The role of flavonoids. Funct. Plant Biol. 2009, 36, 613–620. [Google Scholar] [CrossRef]
- Akhtar, M.S.; Goldschmidt, E.E.; John, I.; Rodoni, S.; Matile, P.; Grierson, D. Altered patterns of senescence and ripening in gf, a stay-green mutant of tomato (Lycopersicon esculentum Mill.). J. Exp. Bot. 1999, 50, 1115–1122. [Google Scholar] [CrossRef]
- Almeida, J.; Azevedo, M.D.S.; Spicher, L.; Glauser, G.; vom Dorp, K.; Guyer, L.; del Valle Carranza, A.; Asis, R.; de Souza, A.P. Down-regulation of tomato PHYTOL KINASE strongly impairs tocopherol biosynthesis and affects prenyllipid metabolism in an organ-specific manner. J. Exp. Bot. 2016, 67, 919–934. [Google Scholar] [CrossRef] [Green Version]
- Lupi, A.C.D.; Lira, B.S.; Gramegna, G.; Trench, B.; Alves, F.R.R.; Demarco, D.; Peres, L.E.P.; Purgatto, E.; Freschi, L.; Rossi, M. Solanum lycopersicum GOLDEN 2-LIKE 2 transcription factor affects fruit quality in a light- and auxin-dependent manner. PLoS ONE 2019, 14, e0212224. [Google Scholar] [CrossRef] [Green Version]
- Lin, C.Y.; Lee, C.H.; Chang, Y.W.; Wang, H.M.; Chen, C.Y.; Chen, Y.H. Pheophytin a inhibits inflammation via suppression of LPS-induced nitric oxide synthase-2, prostaglandin E2, and interleukin-1β of macrophages. Int. J. Mol. 2014, 15, 22819–22834. [Google Scholar] [CrossRef] [Green Version]
- Mishra, V.K.; Bachet, R.K.; Husen, A. Medicinal uses of chlorophyll: A critical overview. In Chlorophyll: Structure, Function and Medicinal Uses; Le, H., Salcedo, E., Eds.; Nova Science Publishers, Inc.: Hauppauge, NY, USA, 2011. [Google Scholar]
- Brigelius-Flohé, R.; Kelly, F.J.; Salonen, J.T.; Neuzil, J.; Zingg, J.M.; Azzi, A. The European perspective on vitamin E: Current knowledge and future research. Am. J. Clin. Nutr. 2002, 76, 703–716. [Google Scholar] [CrossRef] [PubMed]
- Niki, E.; Traber, M.G. History of vitamin E. Ann. Nutr. Metab. 2012, 61, 207–212. [Google Scholar] [CrossRef] [PubMed]








| Class of Material | Class of Variation | Name | Genetic Symbol | Fruit Color |
|---|---|---|---|---|
| Wild-type | - | San Marzano | SM | Red |
| San Marzano fruit variants | Chlorophyll | green flesh | gf | Muddy brown |
| Carotenoids | yellow flesh | r | Yellow | |
| Flavonoids | colorless fruit epidermis | y | Pink | |
| Double mutants | yellow flesh + green flesh | r_gf | Light yellow | |
| - | colorless fruit epidermis + green flesh | y_gf | Wine-coloured | |
| - | colorless fruit epidermis + yellow flesh | y_r | Green |
| Metabolomics Fraction | Metabolic Class | Abbreviation | No. of Compounds | No. of Differentially Accumulated Compounds Over SM | |||||
|---|---|---|---|---|---|---|---|---|---|
| gf | r | y | r_gf | y_gf | y_r | ||||
| Volatile | Benzenoids | B | 8 | 2 | 2 | 3 | 2 | 3 | |
| Branched-chain amino acid derivatives | BCAA | 10 | 3 | 3 | 2 | 4 | 1 | ||
| Apocarotenoids | C | 9 | 3 | 5 | 3 | 6 | 3 | 5 | |
| Esters | E | 2 | |||||||
| Fatty acid derivatives | L | 24 | 3 | 4 | 6 | 1 | 4 | ||
| Others | Phe, S, T, No ID * | 15 | 6 | 4 | 8 | 4 | 6 | ||
| Total | VOCs | 68 | 17 | 18 | 22 | 17 | 3 | 19 | |
| Non-polar | Carotenoids | CAR | 15 | 2 | 5 | 3 | 7 | 5 | 7 |
| Chlorophylls | CHL | 8 | 2 | 1 | 2 | 3 | |||
| Fatty acids | FA | 14 | 1 | 1 | |||||
| Phospholipids | PHO | 1 | 1 | 1 | |||||
| Quinones | QUI | 6 | 1 | 2 | 2 | 3 | |||
| Tocopherols | TOC | 5 | 1 | 1 | 1 | 1 | 2 | ||
| Others | STE, No ID * | 5 | 1 | 1 | 1 | ||||
| Total | NP | 54 | 8 | 10 | 5 | 13 | 10 | 12 | |
| Polar | Amino acids | AA | 19 | 2 | 6 | 3 | 4 | 3 | 6 |
| Acids | AC | 17 | 5 | 5 | 6 | 5 | 4 | 4 | |
| Amines | AM | 4 | 1 | 1 | 1 | ||||
| Sugars and polyols | SAP | 15 | 3 | 3 | 3 | 2 | 4 | 2 | |
| Alkaloids | ALK | 11 | 1 | 4 | 2 | 3 | 1 | 2 | |
| Phenylpropanoids | PHE | 55 | 6 | 13 | 24 | 18 | 18 | 16 | |
| Vitamins | VIT | 3 | 1 | 2 | 1 | ||||
| Others | A, NU, LI, No ID * | 5 | 1 | 2 | 1 | ||||
| Total | P | 129 | 18 | 33 | 40 | 33 | 32 | 33 | |
| Gran total | 251 | 43 | 61 | 67 | 63 | 45 | 64 | ||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dono, G.; Rambla, J.L.; Frusciante, S.; Granell, A.; Diretto, G.; Mazzucato, A. Color Mutations Alter the Biochemical Composition in the San Marzano Tomato Fruit. Metabolites 2020, 10, 110. https://doi.org/10.3390/metabo10030110
Dono G, Rambla JL, Frusciante S, Granell A, Diretto G, Mazzucato A. Color Mutations Alter the Biochemical Composition in the San Marzano Tomato Fruit. Metabolites. 2020; 10(3):110. https://doi.org/10.3390/metabo10030110
Chicago/Turabian StyleDono, Gabriella, Jose Luis Rambla, Sarah Frusciante, Antonio Granell, Gianfranco Diretto, and Andrea Mazzucato. 2020. "Color Mutations Alter the Biochemical Composition in the San Marzano Tomato Fruit" Metabolites 10, no. 3: 110. https://doi.org/10.3390/metabo10030110
APA StyleDono, G., Rambla, J. L., Frusciante, S., Granell, A., Diretto, G., & Mazzucato, A. (2020). Color Mutations Alter the Biochemical Composition in the San Marzano Tomato Fruit. Metabolites, 10(3), 110. https://doi.org/10.3390/metabo10030110