Comparative Untargeted Metabolomics Analysis of the Psychostimulants 3,4-Methylenedioxy-Methamphetamine (MDMA), Amphetamine, and the Novel Psychoactive Substance Mephedrone after Controlled Drug Administration to Humans
Abstract
:1. Introduction
2. Results
2.1. Analytical and Data Processing Procedures of Samples from Controlled Clinical Studies
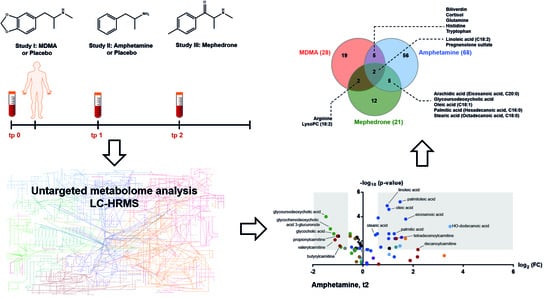
2.2. Effects of MDMA on the Human Metabolome
2.3. Effects of Amphetamine on the Human Metabolome
2.4. Effects of Mephedrone on the Human Metabolome
2.5. Comparative Analysis of MDMA, Amphetamine and Mephedrone Effects
3. Discussion
3.1. Energy Metabolism
3.1.1. Fatty Acids
3.1.2. Acyl Carnitines
3.1.3. Bile Acids
3.2. Steroid Metabolism
4. Materials and Methods
4.1. Clinical Studies
4.1.1. MDMA Study
4.1.2. Amphetamine Study
4.1.3. Mephedrone Study
4.2. Chemicals and Reagents
4.3. Sample Preparation
4.4. LC-HRMS Analysis
4.5. Quality Control
4.6. Data Pre-Processing and Identification
4.7. Statistical Evaluation
4.7.1. Amphetamine
4.7.2. Mephedrone
4.8. Comparative Analysis of MDMA, Amphetamine, and Mephedrone
4.8.1. Comparison of Identified Compounds
4.8.2. Pathway Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- EMCDDA. European Drug Report 2019. Available online: https://www.emcdda.europa.eu/publications/edr/trends-developments/2019 (accessed on 15 June 2020).
- Liechti, M. Novel psychoactive substances (designer drugs): Overview and pharmacology of modulators of monoamine signaling. Swiss Med. Wkly. 2015, 145, w14043. [Google Scholar] [CrossRef] [PubMed]
- Hysek, C.M.; Schmid, Y.; Simmler, L.; Domes, G.; Heinrichs, M.; Eisenegger, C.; Preller, K.H.; Quednow, B.B.; Liechti, M.E. MDMA enhances emotional empathy and prosocial behavior. Soc. Cogn. Affect. Neurosci. 2013, 9, 1645–1652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carvalho, M.; Carmo, H.; Costa, V.M.; Capela, J.P.; Pontes, H.; Remiao, F.; Carvalho, F.; Bastos, M.D.L. Toxicity of amphetamines: An update. Arch. Toxicol. 2012, 86, 1167–1231. [Google Scholar] [CrossRef] [PubMed]
- Holze, F.; Vizeli, P.; Müller, F.; Ley, L.; Duerig, R.; Varghese, N.; Eckert, A.; Borgwardt, S.; Liechti, M.E. Distinct acute effects of LSD, MDMA, and d-amphetamine in healthy subjects. Neuropsychopharmacology 2019, 45, 462–471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kehr, J.; Ichinose, F.; Yoshitake, S.; Goiny, M.; Sievertsson, T.; Nyberg, F.; Yoshitake, T. Mephedrone, compared with MDMA (ecstasy) and amphetamine, rapidly increases both dopamine and 5-HT levels in nucleus accumbens of awake rats. Br. J. Pharmacol. 2011, 164, 1949–1958. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schifano, F.; Mapping, P.W.; Albanese, A.; Fergus, S.; Stair, J.L.; DeLuca, P.; Corazza, O.; Davey, Z.; Corkery, J.M.; Siemann, H.; et al. Mephedrone (4-methylmethcathinone; ‘meow meow’): Chemical, pharmacological and clinical issues. Psychopharmacology 2010, 214, 593–602. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dargan, P.I.; Sedefov, R.; Gallegos, A.; Wood, D.M. The pharmacology and toxicology of the synthetic cathinone mephedrone (4-methylmethcathinone). Drug Test. Anal. 2011, 3, 454–463. [Google Scholar] [CrossRef]
- Hysek, C.M.; Simmler, L.; Nicola, V.G.; Vischer, N.; Donzelli, M.; Krähenbuhl, S.; Grouzmann, E.; Huwyler, J.; Hoener, M.C.; Liechti, M. Duloxetine Inhibits Effects of MDMA (“Ecstasy”) In Vitro and in Humans in a Randomized Placebo-Controlled Laboratory Study. PLoS ONE 2012, 7, e36476. [Google Scholar] [CrossRef] [Green Version]
- Kirkpatrick, M.G.; Francis, S.M.; Lee, R.; De Wit, H.; Jacob, S. Plasma oxytocin concentrations following MDMA or intranasal oxytocin in humans. Psychoneuroendocrinology 2014, 46, 23–31. [Google Scholar] [CrossRef]
- Simmler, L.; Buser, T.; Donzelli, M.; Schramm, Y.; Dieu, L.-H.; Huwyler, J.; Chaboz, S.; Hoener, M.C.; Liechti, M. Pharmacological characterization of designer cathinones in vitro. Br. J. Pharmacol. 2012, 168, 458–470. [Google Scholar] [CrossRef] [Green Version]
- Steuer, A.E.; Brockbals, L.; Kraemer, T. Metabolomic Strategies in Biomarker Research-New Approach for Indirect Identification of Drug Consumption and Sample Manipulation in Clinical and Forensic Toxicology? Front. Chem. 2019, 7, 319. [Google Scholar] [CrossRef] [PubMed]
- Simmler, L.; Rickli, A.; Hoener, M.C.; Liechti, M.E. Monoamine transporter and receptor interaction profiles of a new series of designer cathinones. Neuropharmacology 2014, 79, 152–160. [Google Scholar] [CrossRef] [PubMed]
- Rickli, A.; Hoener, M.C.; Liechti, M.E. Monoamine transporter and receptor interaction profiles of novel psychoactive substances: Para-halogenated amphetamines and pyrovalerone cathinones. Eur. Neuropsychopharmacol. 2015, 25, 365–376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luethi, D.; Kolaczynska, K.E.; Docci, L.; Krähenbühl, S.; Hoener, M.C.; Liechti, M.E. Pharmacological profile of mephedrone analogs and related new psychoactive substances. Neuropharmacology 2018, 134, 4–12. [Google Scholar] [CrossRef] [Green Version]
- Olesti, E.; De Toma, I.; Ramaekers, J.G.; Brunt, T.M.; Carbó, M.; Fernandez-Aviles, C.; Robledo, P.; Farré, M.; Dierssen, M.; Pozo, O.J.; et al. Metabolomics predicts the pharmacological profile of new psychoactive substances. J. Psychopharmacol. 2019, 33, 347–354. [Google Scholar] [CrossRef]
- Bijlsma, L.; Gil Solsona, R.; Hernandez, F.; Sancho, J.V. What about the herb? A new metabolomics approach for synthetic cannabinoid drug testing. Anal. Bioanal. Chem. 2018, 410, 5107–5112. [Google Scholar] [CrossRef]
- Zheng, T.; Liu, L.; Shi, J.; Yu, X.; Xiao, W.; Sun, R.; Zhou, Y.; Aa, J.; Wang, G. The metabolic impact of methamphetamine on the systemic metabolism of rats and potential markers of methamphetamine abuse. Mol. BioSyst. 2014, 10, 1968–1977. [Google Scholar] [CrossRef]
- Dallmann, R.; Viola, A.U.; Tarokh, L.; Cajochen, C.; Brown, S.A. The human circadian metabolome. Proc. Natl. Acad. Sci. USA 2012, 109, 2625–2629. [Google Scholar] [CrossRef] [Green Version]
- O’Sullivan, A.; Gibney, M.J.; Brennan, L. Dietary intake patterns are reflected in metabolomic profiles: Potential role in dietary assessment studies. Am. J. Clin. Nutr. 2011, 93, 314–321. [Google Scholar] [CrossRef] [Green Version]
- Manier, S.K.; Wagmann, L.; Flockerzi, V.; Meyer, M.R. Toxicometabolomics of the new psychoactive substances α-PBP and α-PEP studied in HepaRG cell incubates by means of untargeted metabolomics revealed unexpected amino acid adducts. Arch. Toxicol. 2020, 94, 2047–2059. [Google Scholar] [CrossRef] [Green Version]
- Nielsen, K.L.; Telving, R.; Andreasen, M.; Hasselstrøm, J.B.; Johannsen, M. A Metabolomics Study of Retrospective Forensic Data from Whole Blood Samples of Humans Exposed to 3,4-Methylenedioxymethamphetamine: A New Approach for Identifying Drug Metabolites and Changes in Metabolism Related to Drug Consumption. J. Proteome Res. 2016, 15, 619–627. [Google Scholar] [CrossRef] [PubMed]
- Boxler, M.I.; Liechti, M.E.; Schmid, Y.; Kraemer, T.; Steuer, A.E. First Time View on Human Metabolome Changes after a Single Intake of 3,4-Methylenedioxymethamphetamine in Healthy Placebo-Controlled Subjects. J. Proteome Res. 2017, 16, 3310–3320. [Google Scholar] [CrossRef]
- Boxler, M.I.; Streun, G.L.; Liechti, M.E.; Schmid, Y.; Kraemer, T.; Steuer, A.E. Human Metabolome Changes after a Single Dose of 3,4-Methylenedioxymethamphetamine (MDMA) with Special Focus on Steroid Metabolism and Inflammation Processes. J. Proteome Res. 2018, 17, 2900–2907. [Google Scholar] [CrossRef] [PubMed]
- Patkar, A.A.; Rozen, S.; Mannelli, P.; Matson, W.; Pae, C.-U.; Krishnan, K.R.; Kaddurah-Daouk, R.; Rozen, S.G. Alterations in tryptophan and purine metabolism in cocaine addiction: A metabolomic study. Psychopharmacology 2009, 206, 479–489. [Google Scholar] [CrossRef]
- Costa, T.B.B.C.; Lacerda, A.L.; Mas, C.D.; Brietzke, E.; Pontes, J.G.M.; Marins, L.A.N.; Martins, L.G.; Nunes, M.V.; Pedrini, M.; Carvalho, M.S.C.; et al. Insights into the Effects of Crack Abuse on the Human Metabolome Using a NMR Approach. J. Proteome Res. 2019, 18, 341–348. [Google Scholar] [CrossRef] [PubMed]
- Steuer, A.E.; Raeber, J.; Steuer, C.; Boxler, M.I.; Dornbierer, D.A.; Bosch, O.G.; Quednow, B.B.; Seifritz, E.; Kraemer, T. Identification of new urinary gamma-hydroxybutyric acid markers applying untargeted metabolomics analysis following placebo-controlled administration to humans. Drug Test. Anal. 2019, 11, 813–823. [Google Scholar] [CrossRef]
- Piper, T.; Mehling, L.-M.; Spottke, A.; Heidbreder, A.; Young, P.; Madea, B.; Hess, C.; Schänzer, W.; Thevis, M. Potential of GHB phase-II-metabolites to complement current approaches in GHB post administration detection. Forensic Sci. Int. 2017, 279, 157–164. [Google Scholar] [CrossRef]
- Palomino-Schätzlein, M.; Wang, Y.; Brailsford, A.D.; Parella, T.; Cowan, A.D.; Legido-Quigley, C.; Pérez-Trujillo, M. Direct Monitoring of Exogenous γ-Hydroxybutyric Acid in Body Fluids by NMR Spectroscopy. Anal. Chem. 2017, 89, 8343–8350. [Google Scholar] [CrossRef] [Green Version]
- Schmid, Y.; Rickli, A.; Schaffner, A.; Duthaler, U.; Grouzmann, E.; Hysek, C.M.; Liechti, M.E. Interactions between Bupropion and 3,4-Methylenedioxymethamphetamine in Healthy Subjects. J. Pharmacol. Exp. Ther. 2015, 353, 102–111. [Google Scholar] [CrossRef]
- Boxler, M.I.; Schneider, T.D.; Kraemer, T.; Steuer, A.E. Analytical considerations for (un)-targeted metabolomic studies with special focus on forensic applications. Drug Test. Anal. 2019, 11, 678–696. [Google Scholar] [CrossRef]
- Sumner, L.W.; Amberg, A.; Barrett, D.A.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.-M.; Fiehn, O.; Goodacre, R.; Griffin, J.; et al. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef] [Green Version]
- Chong, J.; Wishart, D.S.; Xia, J. Using MetaboAnalyst 4.0 for Comprehensive and Integrative Metabolomics Data Analysis. Curr. Protoc. Bioinform. 2019, 68, e86. [Google Scholar] [CrossRef] [PubMed]
- Barri, T.; Dragsted, L.O. UPLC-ESI-QTOF/MS and multivariate data analysis for blood plasma and serum metabolomics: Effect of experimental artefacts and anticoagulant. Anal. Chim. Acta 2013, 768, 118–128. [Google Scholar] [CrossRef]
- Li, S.; Park, Y.H.; Duraisingham, S.; Strobel, F.H.; Khan, N.; Soltow, Q.A.; Jones, D.P.; Pulendran, B. Predicting Network Activity from High Throughput Metabolomics. PLoS Comput. Boil. 2013, 9, e1003123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xia, J.; Wishart, D.S. MSEA: A web-based tool to identify biologically meaningful patterns in quantitative metabolomic data. Nucleic Acids Res. 2010, 38, W71–W77. [Google Scholar] [CrossRef] [Green Version]
- Computational Methods and Data Analysis for Metabolomics; Li, S. (Ed.) Humana Press: Totowa, NJ, USA, 2020. [Google Scholar]
- Pękala, J.W.; Patkowska-Sokoła, B.; Bodkowski, R.; Jamroz, D.; Nowakowski, P.; Lochyński, S.; Librowski, T. L-carnitine--metabolic functions and meaning in humans life. Curr. Drug Metab. 2011, 12, 667–678. [Google Scholar] [CrossRef]
- Chiang, J.Y., III. Bile acids and nuclear receptors. Am. J. Physiol. Liver Physiol. 2003, 284, G349–G356. [Google Scholar] [CrossRef] [Green Version]
- Li, T.; Chiang, J.Y. Bile acid signaling in metabolic disease and drug therapy. Pharmacol. Rev. 2014, 66, 948–983. [Google Scholar] [CrossRef] [Green Version]
- Comai, K.; Triscari, J.; Sullivan, A.C. Comparative effects of amphetamine and fenfluramine on lipid biosynthesis and absorption in the rat. Biochem. Pharmacol. 1978, 27, 1987–1994. [Google Scholar] [CrossRef]
- Zhang, M.; Lv, D.; Zhou, W.; Ji, L.; Zhou, B.; Chen, H.; Gu, Y.; Zhao, J.; He, J. The levels of triglyceride and total cholesterol in methamphetamine dependence. Medicine 2017, 96, e6631. [Google Scholar] [CrossRef] [PubMed]
- Calder, P.C. Functional Roles of Fatty Acids and Their Effects on Human Health. J. Parenter. Enter. Nutr. 2015, 39, 18S–32S. [Google Scholar] [CrossRef] [PubMed]
- Kingsbury, K.J.; Paul, S.; Crossley, A.; Morgan, D.M. The fatty acid composition of human depot fat. Biochem. J. 1961, 78, 541–550. [Google Scholar] [CrossRef] [PubMed]
- Aruoma, O.I.; Gootveld, M.; Bahorun, T. Free radicals in biology and medicine: From inflammation to biotechnology. BioFactors 2006, 27, 1–3. [Google Scholar] [CrossRef]
- Quinton, M.S.; Yamamoto, B.K. Causes and consequences of methamphetamine and MDMA toxicity. AAPS J. 2006, 8, E337. [Google Scholar] [CrossRef] [Green Version]
- Perrine, S.A.; Michaels, M.S.; Ghoddoussi, F.; Hyde, E.M.; Tancer, M.E.; Galloway, M.P. Cardiac effects of MDMA on the metabolic profile determined with1H-magnetic resonance spectroscopy in the rat. NMR Biomed. 2009, 22, 419–425. [Google Scholar] [CrossRef] [Green Version]
- Vanaveski, T.; Narvik, J.; Innos, J.; Philips, M.-A.; Ottas, A.; Plaas, M.; Haring, L.; Zilmer, M.; Vasar, E. Repeated Administration of D-Amphetamine Induces Distinct Alterations in Behavior and Metabolite Levels in 129Sv and Bl6 Mouse Strains. Front. Mol. Neurosci. 2018, 12, 399. [Google Scholar] [CrossRef]
- Famularo, G.; Simone, C.; Trinchieri, V.; Mosca, L. Carnitines and Its Congeners: A Metabolic Pathway to the Regulation of Immune Response and Inflammation. Ann. N. Y. Acad. Sci. 2004, 1033, 132–138. [Google Scholar] [CrossRef]
- Strajhar, P.; Vizeli, P.; Patt, M.; Dolder, P.C.; Kratschmar, D.V.; Liechti, M.E.; Odermatt, W.M.A.A. Effects of lisdexamfetamine on plasma steroid concentrations compared with d-amphetamine in healthy subjects: A randomized, double-blind, placebo-controlled study. J. Steroid Biochem. Mol. Boil. 2019, 186, 212–225. [Google Scholar] [CrossRef] [Green Version]
- Parrott, A.C. Oxytocin, cortisol and 3,4-methylenedioxymethamphetamine. Behav. Pharmacol. 2016, 27, 649–658. [Google Scholar] [CrossRef] [Green Version]
- Hysek, C.M.; Simmler, L.; Schillinger, N.; Meyer, N.; Schmid, Y.; Donzelli, M.; Grouzmann, E.; Liechti, M.E. Pharmacokinetic and pharmacodynamic effects of methylphenidate and MDMA administered alone or in combination. Int. J. Neuropsychopharmacol. 2014, 17, 371–381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seibert, J.; Hysek, C.M.; Penno, C.A.; Schmid, Y.; Kratschmar, D.V.; Liechti, M.E.; Odermatt, W.M.A.A. Acute Effects of 3,4-Methylenedioxymethamphetamine and Methylphenidate on Circulating Steroid Levels in Healthy Subjects. Neuroendocrinology 2014, 100, 17–25. [Google Scholar] [CrossRef] [Green Version]
- Lefebvre, H.; Contesse, V.; Delarue, C.; Feuilloley, M.; Héry, F.; Grise, P.; Raynaud, G.; Verhofstad, A.; Wolf, L.; Vaudry, H. Serotonin-induced stimulation of cortisol secretion from human adrenocortical tissue is mediated through activation of a serotonin4 receptor subtype. Neuroscience 1992, 47, 999–1007. [Google Scholar] [CrossRef]
- Zorumski, C.F.; Paul, S.M.; Izumi, Y.; Covey, D.F.; Mennerick, S. Neurosteroids, stress and depression: Potential therapeutic opportunities. Neurosci. Biobehav. Rev. 2013, 37, 109–122. [Google Scholar] [CrossRef] [Green Version]
- Vallée, M. Neurosteroids and potential therapeutics: Focus on pregnenolone. J. Steroid Biochem. Mol. Boil. 2016, 160, 78–87. [Google Scholar] [CrossRef]
- Semeniuk, T.; Jhangri, G.S.; Le Mellédo, J.-M. Neuroactive Steroid Levels in Patients With Generalized Anxiety Disorder. J. Neuropsychiatry Clin. Neurosci. 2001, 13, 396–398. [Google Scholar] [CrossRef] [PubMed]
- Heydari, B.; Le Mellédo, J.-M. Low pregnenolone sulphate plasma concentrations in patients with generalized social phobia. Psychol. Med. 2002, 32, 929–933. [Google Scholar] [CrossRef] [Green Version]
- Czerwinska, J.; Jang, M.; Costa, C.; Parkin, M.C.; George, C.; Kicman, A.T.; Bailey, M.; Dargan, P.I.; Abbate, V. Detection of mephedrone and its metabolites in fingerprints from a controlled human administration study by liquid chromatography-tandem mass spectrometry and paper spray-mass spectrometry. Analyst 2020, 145, 3038–3048. [Google Scholar] [CrossRef]
- Guijas, C.; Montenegro-Burke, J.R.; Domingo-Almenara, X.; Palermo, A.; Warth, B.; Hermann, G.; Koellensperger, G.; Huan, T.; Uritboonthai, W.; Aisporna, A.E.; et al. METLIN: A Technology Platform for Identifying Knowns and Unknowns. Anal. Chem. 2018, 90, 3156–3164. [Google Scholar] [CrossRef] [Green Version]
- Wishart, D.S.; Tzur, D.; Knox, C.; Eisner, R.; Guo, A.C.; Young, N.; Cheng, D.; Jewell, K.; Arndt, D.; Sawhney, S.; et al. HMDB: The Human Metabolome Database. Nucleic Acids Res. 2007, 35, D521–D526. [Google Scholar] [CrossRef]
- Linstrom, P.; Mallard, W.G. NIST Chemistry WebBook; NIST Standard Reference Database No. 692001. Available online: https://webbook.nist.gov/chemistry/ (accessed on 26 July 2020).
- Kind, T.; Liu, K.-H.; Lee, D.Y.; DeFelice, B.; Meissen, J.K.; Fiehn, O. LipidBlast in silico tandem mass spectrometry database for lipid identification. Nat. Methods 2013, 10, 755–758. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chong, J.; Soufan, O.; Li, C.; Caraus, I.; Li, S.; Bourque, G.; Wishart, D.S.; Xia, J. MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018, 46, W486–W494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heberle, H.; Meirelles, G.V.; Da Silva, F.R.; Telles, G.P.; Minghim, R. InteractiVenn: A web-based tool for the analysis of sets through Venn diagrams. BMC Bioinform. 2015, 16, 169. [Google Scholar] [CrossRef] [PubMed]






© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Steuer, A.E.; Kaelin, D.; Boxler, M.I.; Eisenbeiss, L.; Holze, F.; Vizeli, P.; Czerwinska, J.; Dargan, P.I.; Abbate, V.; Liechti, M.E.; et al. Comparative Untargeted Metabolomics Analysis of the Psychostimulants 3,4-Methylenedioxy-Methamphetamine (MDMA), Amphetamine, and the Novel Psychoactive Substance Mephedrone after Controlled Drug Administration to Humans. Metabolites 2020, 10, 306. https://doi.org/10.3390/metabo10080306
Steuer AE, Kaelin D, Boxler MI, Eisenbeiss L, Holze F, Vizeli P, Czerwinska J, Dargan PI, Abbate V, Liechti ME, et al. Comparative Untargeted Metabolomics Analysis of the Psychostimulants 3,4-Methylenedioxy-Methamphetamine (MDMA), Amphetamine, and the Novel Psychoactive Substance Mephedrone after Controlled Drug Administration to Humans. Metabolites. 2020; 10(8):306. https://doi.org/10.3390/metabo10080306
Chicago/Turabian StyleSteuer, Andrea E., Daria Kaelin, Martina I. Boxler, Lisa Eisenbeiss, Friederike Holze, Patrick Vizeli, Joanna Czerwinska, Paul I. Dargan, Vincenzo Abbate, Matthias E. Liechti, and et al. 2020. "Comparative Untargeted Metabolomics Analysis of the Psychostimulants 3,4-Methylenedioxy-Methamphetamine (MDMA), Amphetamine, and the Novel Psychoactive Substance Mephedrone after Controlled Drug Administration to Humans" Metabolites 10, no. 8: 306. https://doi.org/10.3390/metabo10080306
APA StyleSteuer, A. E., Kaelin, D., Boxler, M. I., Eisenbeiss, L., Holze, F., Vizeli, P., Czerwinska, J., Dargan, P. I., Abbate, V., Liechti, M. E., & Kraemer, T. (2020). Comparative Untargeted Metabolomics Analysis of the Psychostimulants 3,4-Methylenedioxy-Methamphetamine (MDMA), Amphetamine, and the Novel Psychoactive Substance Mephedrone after Controlled Drug Administration to Humans. Metabolites, 10(8), 306. https://doi.org/10.3390/metabo10080306