Comparative Lipidomics of Different Yeast Species Associated to Drosophila suzukii
Abstract
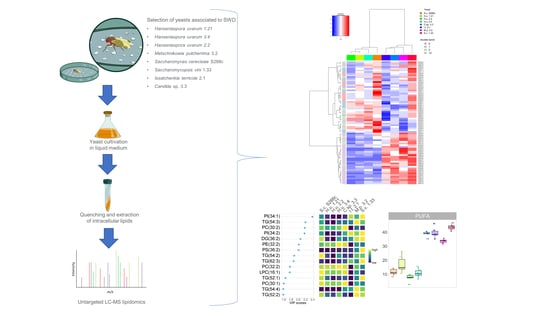
:1. Introduction
2. Results and Discussion
2.1. Compound Annotation and Differences in the Lipid Profiles
2.2. Compound Classes Responsible for Discrimination between Yeast Species
2.3. Differences in the Lipid Profile of GP, DG and TG
2.4. Differences in the Lipid Profile of FA, Ceramides, LCB and Sterols
2.5. Relationship between the Selected Yeasts and SWD
3. Materials and Methods
3.1. Chemicals and Growth Media
3.2. Yeasts Cultivation and Lipid Extraction
3.3. Chromatographic and Mass Spectrometric Conditions
3.4. Data Processing and Statistics
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hauser, M. A historic account of the invasion of Drosophila suzukii (Matsumura) (Diptera: Drosophilidae) in the continental United States, with remarks on their identification. Pest Manag. Sci. 2011, 67, 1352–1357. [Google Scholar] [CrossRef] [PubMed]
- Cini, A.; Anfora, G.; Escudero-Colomar, L.A.; Grassi, A.; Santosuosso, U.; Seljak, G.; Papini, A. Tracking the invasion of the alien fruit pest Drosophila suzukii in Europe. J. Pest Sci. 2014, 87, 559–566. [Google Scholar] [CrossRef]
- De Ros, G.; Conci, S.; Pantezzi, T.; Savini, G. The economic impact of invasive pest Drosophila suzukii on berry production in the Province of Trento, Italy. J. Berry Res. 2015, 5, 89–96. [Google Scholar] [CrossRef] [Green Version]
- Dos Santos, L.A.; Mendes, M.F.; Krüger, A.P.; Blauth, M.L.; Gottschalk, M.S.; Garcia, F.R.M. Global potential distribution of Drosophila suzukii (Diptera, Drosophilidae). PLoS ONE 2017, 12, e0174318. [Google Scholar] [CrossRef] [Green Version]
- Hamby, K.A.; Hernández, A.; Boundy-Mills, K.; Zalom, F.G. Associations of yeasts with spotted-wing Drosophila (Drosophila suzukii; Diptera: Drosophilidae) in cherries and raspberries. Appl. Environ. Microbiol. 2012, 78, 4869–4873. [Google Scholar] [CrossRef] [Green Version]
- Bellutti, N.; Gallmetzer, A.; Innerebner, G.; Schmidt, S.; Zelger, R.; Koschier, E.H. Dietary yeast affects preference and performance in Drosophila suzukii. J. Pest. Sci. 2018, 91, 651–660. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lewis, M.T.; Hamby, K.A. Differential Impacts of Yeasts on Feeding Behavior and Development in Larval Drosophila suzukii (Diptera: Drosophilidae). Sci. Rep. 2019, 9, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Iglesias, L.E.; Nyoike, T.W.; Liburd, O.E. Effect of Trap Design, Bait Type, and Age on Captures of Drosophila suzukii (Diptera: Drosophilidae) in Berry Crops. J. Econ. Entomol. 2014, 107, 1508–1518. [Google Scholar] [CrossRef] [Green Version]
- Hamby, K.A.; Becher, P.G. Current knowledge of interactions between Drosophila suzukii and microbes, and their potential utility for pest management. J. Pest. Sci. 2016, 89, 621–630. [Google Scholar] [CrossRef]
- Knight, A.L.; Basoalto, E.; Yee, W.; Hilton, R.; Kurtzman, C.P. Adding yeasts with sugar to increase the number of effective insecticide classes to manage Drosophila suzukii (Matsumura) (Diptera: Drosophilidae) in cherry. Pest. Manag. Sci. 2016, 72, 1482–1490. [Google Scholar] [CrossRef]
- Mori, B.A.; Whitener, A.B.; Leinweber, Y.; Revadi, S.; Beers, E.H.; Witzgall, P.; Becher, P.G. Enhanced yeast feeding following mating facilitates control of the invasive fruit pest Drosophila suzukii. J. Appl. Ecol. 2017, 54, 170–177. [Google Scholar] [CrossRef]
- Becher, P.G.; Flick, G.; Rozpedowska, E.; Schmidt, A.; Hagman, A.; Lebreton, S.; Larsson, M.C.; Hansson, B.S.; Piškur, J.; Witzgall, P.; et al. Yeast, not fruit volatiles mediate Drosophila melanogaster attraction, oviposition and development. Funct. Ecol. 2012, 26, 822–828. [Google Scholar] [CrossRef]
- De Camargo, R.; Phaff, H.J. Yeasts occurring in Drosophila flies and in fermenting tomato fruits in Northern California. J. Food Sci. 1957, 22, 367–372. [Google Scholar] [CrossRef]
- Carvalho, M.; Sampaio, J.L.; Palm, W.; Brankatschk, M.; Eaton, S.; Shevchenko, A. Effects of diet and development on the Drosophila lipidome. Mol. Syst. Biol. 2012, 8, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Yamada, R.; Deshpande, S.A.; Bruce, K.D.; Mak, E.M.; Ja, W.W. Microbes Promote Amino Acid Harvest to Rescue Undernutrition in Drosophila. Cell Rep. 2015, 10, 865–872. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Steck, K.; Walker, S.J.; Itskov, P.M.; Baltazar, C.; Moreira, J.M.; Ribeiro, C. Internal amino acid state modulates yeast taste neurons to support protein homeostasis in Drosophila. Elife 2018, 7, 1–29. [Google Scholar] [CrossRef]
- Bing, X.L.; Gerlach, J.; Loeb, G.; Buchon, N. Nutrient-dependent impact of microbes on Drosophila suzukii development. MBio 2018, 9, e02199-17. [Google Scholar] [CrossRef] [Green Version]
- Madden, A.A.; Epps, M.J.; Fukami, T.; Irwin, R.E.; Sheppard, J.; Sorger, D.M.; Dunn, R.R. The ecology of insect—Yeast relationships and its relevance to human industry. Proc. R. Soc. B Biol. Sci. 2018, 285, 20172733. [Google Scholar] [CrossRef] [Green Version]
- Scheidler, N.H.; Liu, C.; Hamby, K.A.; Zalom, F.G.; Syed, Z. Volatile codes: Correlation of olfactory signals and reception in Drosophila-yeast chemical communication. Sci. Rep. 2015, 5, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Lasa, R.; Navarro-De-La-Fuente, L.; Gschaedler-Mathis, A.C.; Kirchmayr, M.R.; Williams, T. Yeast species, strains, and growth media mediate attraction of Drosophila suzukii (Diptera: Drosophilidae). Insects 2019, 10, 228. [Google Scholar] [CrossRef] [Green Version]
- Fanson, B.G.; Taylor, P.W. Protein:carbohydrate ratios explain life span patterns found in Queensland fruit fly on diets varying in yeast:sugar ratios. Age 2012, 34, 1361–1368. [Google Scholar] [CrossRef] [Green Version]
- Douglas, A.E. The B vitamin nutrition of insects: The contributions of diet, microbiome and horizontally acquired genes. Curr. Opin. Insect Sci. 2017, 23, 65–69. [Google Scholar] [CrossRef] [PubMed]
- Rudkin, G.T.; Schultz, J. Evolution of nutritional requirements in animals; amino-acids essential for Drosophilia melanogaster. Anat. Rec. 1947, 99, 613. [Google Scholar] [PubMed]
- Royes, W.V.; Robertson, F.W. The nutritional requirements and growth relations of different species of Drosophila. J. Exp. Zool. 1964, 156, 105–135. [Google Scholar] [CrossRef]
- Markow, T.A.; O’Grady, P. Reproductive ecology of Drosophila. Funct. Ecol. 2008, 22, 747–759. [Google Scholar] [CrossRef]
- Fahy, E.; Cotter, D.; Sud, M.; Subramaniam, S. Lipid classification, structures and tools. Biochim. Biophys. Acta-Mol. Cell Biol. Lipids 2011, 1811, 637–647. [Google Scholar] [CrossRef] [PubMed]
- Tuller, G.; Nemec, T.; Hrastnik, C.; Daum, G. Lipid composition of subcellular membranes of an FY1679-derived haploid yeast wild-type strain grown on different carbon sources. Yeast 1999, 15, 1555–1564. [Google Scholar] [CrossRef]
- Tehlivets, O.; Scheuringer, K.; Kohlwein, S.D. Fatty acid synthesis and elongation in yeast. Biochim. Biophys. Acta-Mol. Cell Biol. Lipids 2007, 1771, 255–270. [Google Scholar] [CrossRef]
- Hein, E.M.; Hayen, H. Comparative lipidomic profiling of S. cerevisiae and four other hemiascomycetous yeasts. Metabolites 2012, 2, 254–267. [Google Scholar] [CrossRef] [Green Version]
- Koch, B.; Schmidt, C.; Daum, G. Storage lipids of yeasts: A survey of nonpolar lipid metabolism in Saccharomyces cerevisiae, Pichia pastoris, and Yarrowia lipolytica. FEMS Microbiol. Rev. 2014, 38, 892–915. [Google Scholar] [CrossRef] [Green Version]
- Czabany, T.; Wagner, A.; Zweytick, D.; Lohner, K.; Leitner, E.; Ingolic, E.; Daum, G. Structural and biochemical properties of lipid particles from the yeast Saccharomyces cerevisiae. J. Biol. Chem. 2008, 283, 17065–17074. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cowart, L.A.; Obeid, L.M. Yeast sphingolipids: Recent developments in understanding biosynthesis, regulation, and function. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2007, 1771, 421–431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pina, C.; Santos, C.; Couto, J.A.; Hogg, T. Ethanol tolerance of five non-Saccharomyces wine yeasts in comparison with a strain of Saccharomyces cerevisiae—Influence of different culture conditions. Food Microbiol. 2004, 21, 439–447. [Google Scholar] [CrossRef]
- Kaneko, H.; Hosohara, M.; Tanaka, M.; Itoh, T. Lipid composition of 30 species of yeast. Lipids 1976, 11, 837–844. [Google Scholar] [CrossRef] [PubMed]
- Ejsing, C.S.; Sampaio, J.L.; Surendranath, V.; Duchoslav, E.; Ekroos, K.; Klemm, R.W.; Simons, K.; Shevchenko, A. Global analysis of the yeast lipidome by quantitative shotgun mass spectrometry. Proc. Natl. Acad. Sci. USA 2009, 106, 2136–2141. [Google Scholar] [CrossRef] [Green Version]
- Klug, L.; Daum, G. Yeast lipid metabolism at a glance. FEMS Yeast Res. 2014, 14, 369–388. [Google Scholar] [CrossRef] [Green Version]
- Pires, E.J.; Teixeira, J.A.; Brányik, T.; Vicente, A.A. Yeast: The soul of beer’s aroma—A review of flavour-active esters and higher alcohols produced by the brewing yeast. Appl. Microbiol. Biotechnol. 2014, 98, 1937–1949. [Google Scholar] [CrossRef] [Green Version]
- Rollero, S.; Mouret, J.R.; Sanchez, I.; Camarasa, C.; Ortiz-Julien, A.; Sablayrolles, J.M.; Dequin, S. Key role of lipid management in nitrogen and aroma metabolism in an evolved wine yeast strain. Microb. Cell Fact. 2016, 15, 1–15. [Google Scholar] [CrossRef]
- Aslankoohi, E.; Herrera-Malaver, B.; Rezaei, M.N.; Steensels, J.; Courtin, C.M.; Verstrepen, K.J. Non-conventional yeast strains increase the aroma complexity of bread. PLoS ONE 2016, 11, e0165126. [Google Scholar] [CrossRef] [Green Version]
- Fellman, J.K.; Miller, T.W.; Mattinson, D.S.; Mattheis, J.P. Factors that influence biosynthesis of volatile flavor compounds in apple fruits. HortScience 2000, 35, 1026–1033. [Google Scholar] [CrossRef]
- Song, J.; Bangerth, F. Fatty acids as precursors for aroma volatile biosynthesis in pre-climacteric and climacteric apple fruit. Postharvest Biol. Technol. 2003, 30, 113–121. [Google Scholar] [CrossRef]
- Van Meer, G. Cellular lipidomics. EMBO J. 2005, 24, 3159–3165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wenk, M.R. The emerging field of lipidomics. Nat. Rev. Drug Discov. 2005, 4, 594–610. [Google Scholar] [CrossRef]
- Gaspar, M.L.; Aregullin, M.A.; Jesch, S.A.; Nunez, L.R.; Villa-García, M.; Henry, S.A. The emergence of yeast lipidomics. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2007, 1771, 241–254. [Google Scholar] [CrossRef] [PubMed]
- Watson, A.D. Lipidomics: A global approach to lipid analysis in biological systems. J. Lipid Res. 2006, 47, 2101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wolf, C.; Quinn, P.J. Lipidomics: Practical aspects and applications. Prog. Lipid Res. 2008, 47, 15–36. [Google Scholar] [CrossRef]
- Li, M.; Yang, L.; Bai, Y.; Liu, H. Analytical methods in lipidomics and their applications. Anal. Chem. 2014, 86, 161–175. [Google Scholar] [CrossRef]
- Casanovas, A.; Sprenger, R.R.; Tarasov, K.; Ruckerbauer, D.E.; Hannibal-Bach, H.K.; Zanghellini, J.; Jensen, O.N.; Ejsing, C.S. Quantitative analysis of proteome and lipidome dynamics reveals functional regulation of global lipid metabolism. Chem. Biol. 2015, 22, 412–425. [Google Scholar] [CrossRef] [Green Version]
- Aloklah, B.; Alhajali, A.; Yaziji, S. Identification of some yeasts by fatty acid profiles. Polish J. Microbiol. 2014, 63, 467–472. [Google Scholar] [CrossRef]
- Viljoen, B.C.; Kock, J.L.F.; Lategan, P.M. Fatty acid composition as a guide to the classification of selected genera of yeasts belonging to the endomycetales. J. Gen. Microbiol. 1986, 132, 2397–2400. [Google Scholar] [CrossRef] [Green Version]
- Augustyn, O.P.H.; Ferreira, D.; Kock, J.L.F. Differentiation between Yeast Species, and Strains within a Species, by Cellular Fatty Acid Analysis: 4. Saccharomyces sensu stricto, Hanseniaspora, Saccharomycodes and Wickerhamiella. Syst. Appl. Microbiol. 1991, 14, 324–334. [Google Scholar] [CrossRef]
- Spitaler, U.; Bianchi, F.; Eisenstecken, D.; Castellan, I.; Angeli, S.; Dordevic, N.; Robatscher, P.; Vogel, R.F.; Koschier, E.H.; Schmidt, S. Yeast species affects feeding and fitness of Drosophila suzukii adults. J. Pest Sci. 2020, 93, 1295–1309. [Google Scholar] [CrossRef]
- Jewison, T.; Knox, C.; Neveu, V.; Djoumbou, Y.; Guo, A.C.; Lee, J.; Liu, P.; Mandal, R.; Krishnamurthy, R.; Sinelnikov, I.; et al. YMDB: The yeast metabolome database. Nucleic Acids Res. 2012, 40, D815–D820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chandler, J.A.; Eisen, J.A.; Kopp, A. Yeast communities of diverse Drosophila species: Comparison of two symbiont groups in the same hosts. Appl. Environ. Microbiol. 2012, 87, 559–566. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fountain, M.T.; Bennett, J.; Cobo-Medina, M.; Conde Ruiz, R.; Deakin, G.; Delgado, A.; Harrison, R.; Harrison, N. Alimentary microbes of winter-form Drosophila suzukii. Insect Mol. Biol. 2018, 27, 383–392. [Google Scholar] [CrossRef] [PubMed]
- Crous, P.W.; Gams, W.; Stalpers, J.A.; Robert, V.; Stegehuis, G. MycoBank: An online initiative to launch mycology into the 21st century. Stud. Mycol. 2004, 50, 19–22. [Google Scholar]
- Robert, V.; Stegehuis, G.; Stalpers, J. Mycobank. The MycoBank Engine and Related Databases 2005. Available online: http://www.mycobank.org/defaultinfo.aspx?Page=Home (accessed on 30 January 2020).
- Robert, V.; Vu, D.; Amor, A.B.H.; van de Wiele, N.; Brouwer, C.; Jabas, B.; Szoke, S.; Dridi, A.; Triki, M.; Daoud, S.B.; et al. MycoBank gearing up for new horizons. IMA Fungus 2013, 4, 371–379. [Google Scholar] [CrossRef]
- Rattray, J.B.M.; Schibeci, A.; Kidby, D.K. Lipids of yeasts. Bacteriol. Rev. 1975, 39, 197–231. [Google Scholar] [CrossRef] [Green Version]
- Mishina, M.; Yanagawa, S.; Tanaka, A.; Fukui, S. Effects of chain-length of alkane substrate on fatty acid composition and biosynthetic pathway in some Candida yeasts. Agric. Biol. Chem. 1973, 37, 863–870. [Google Scholar] [CrossRef]
- Řezanka, T.; Sigler, K. Odd-numbered very-long-chain fatty acids from the microbial, animal and plant kingdoms. Prog. Lipid Res. 2009, 48, 206–238. [Google Scholar] [CrossRef]
- Kondo, N.; Ohno, Y.; Yamagata, M.; Obara, T.; Seki, N.; Kitamura, T.; Naganuma, T.; Kihara, A. Identification of the phytosphingosine metabolic pathway leading to odd-numbered fatty acids. Nat. Commun. 2014, 5, 5338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, H.; Kim, H.; Kwon, J.Y.; Seo, J.T.; Shin, D.M.; Moon, S.J. Drosophila Gr64e mediates fatty acid sensing via the phospholipase C pathway. PLoS Genet. 2018, 14, e1007229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Masek, P.; Keene, A.C. Drosophila Fatty Acid Taste Signals through the PLC Pathway in Sugar-Sensing Neurons. PLoS Genet. 2013, 9, e1003710. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yazawa, H.; Iwahashi, H.; Kamisaka, Y.; Kimura, K.; Uemura, H. Production of polyunsaturated fatty acids in yeast Saccharomyces cerevisiae and its relation to alkaline pH tolerance. Yeast 2009, 26, 167–184. [Google Scholar] [CrossRef]
- Rezanka, T. Very-long-chain fatty acids from the animal and plant kingdoms. Prog. Lipid Res. 1989, 28, 147–187. [Google Scholar] [CrossRef]
- Silva-Soares, N.F.; Nogueira-Alves, A.; Beldade, P.; Mirth, C.K. Adaptation to new nutritional environments: Larval performance, foraging decisions, and adult oviposition choices in Drosophila suzukii. BMC Ecol. 2017, 17, 21. [Google Scholar] [CrossRef] [Green Version]
- Anagnostou, C.; Dorsch, M.; Rohlfs, M. Influence of dietary yeasts on Drosophila melanogaster life-history traits. Entomol. Exp. Appl. 2010, 136, 1–11. [Google Scholar] [CrossRef]
- Barupal, D.K.; Fiehn, O. Chemical Similarity Enrichment Analysis (ChemRICH) as alternative to biochemical pathway mapping for metabolomic datasets. Sci. Rep. 2017, 7, 14567. [Google Scholar] [CrossRef]
- Showalter, M.R.; Nonnecke, E.B.; Linderholm, A.L.; Cajka, T.; Sa, M.R.; Lönnerdal, B.; Kenyon, N.J.; Fiehn, O. Obesogenic diets alter metabolism in mice. PLoS ONE 2018, 13, e0190632. [Google Scholar] [CrossRef] [Green Version]
- Tsugawa, H.; Cajka, T.; Kind, T.; Ma, Y.; Higgins, B.; Ikeda, K.; Kanazawa, M.; Vandergheynst, J.; Fiehn, O.; Arita, M. MS-DIAL: Data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat. Methods 2015, 12, 523–526. [Google Scholar] [CrossRef]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.M.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed minimum reporting standards for chemical analysis. Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Züllig, T.; Trötzmüller, M.; Köfeler, H.C. Lipidomics from sample preparation to data analysis: A primer. Anal. Bioanal. Chem. 2020, 412, 2191–2209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Team, R.C. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019. [Google Scholar]
- Haug, K.; Cochrane, K.; Nainala, V.C.; Williams, M.; Chang, J.; Jayaseelan, K.V.; O’Donovan, C. MetaboLights: A resource evolving in response to the needs of its scientific community. Nucleic Acids Res. 2020, 48, D440–D444. [Google Scholar] [CrossRef] [PubMed] [Green Version]





| Yeast Species | Strain | Accession Number * | Abbreviation | OD600 | CDW(mg mL−1 Fermentation Broth) |
|---|---|---|---|---|---|
| Saccharomyces cerevisiae | S288c | - | S.c. S288c | 1.98 ± 0.03 | 1.57 ± 0.18 |
| Hanseniaspora uvarum | LB-NB-1.21 | KP298009 | H.u. 1.21 | 1.86 ± 0.02 | 1.42 ± 0.11 |
| Hanseniaspora uvarum | LB-NB-2.2 | MK567898 | H.u. 2.2 | 1.83 ± 0.05 | 1.48 ± 0.15 |
| Hanseniaspora uvarum | LB-NB-3.4 | MK567905 | H.u. 3.4 | 1.86 ± 0.04 | 1.59 ± 0.06 |
| Issatchenkia/Picchia terricola | LB-NB-2.1 | MK567903 | I.t. 2.1 | 1.90 ± 0.04 | 1.32 ± 0.17 |
| Metschnikowia pulcherrima | LB-NB-3.2 | KP298012 | M.p. 3.2 | 2.02 ± 0.05 | 1.94 ± 0.15 |
| Saccharomycopsis vini | LB-NB-1.33 | KP298011 | S.v. 1.33 | 1.78 ± 0.10 | 1.63 ± 0.13 |
| Candida sp. | LB-NB-3.3 | KP298013 | C.sp. 3.3 | 2.04 ± 0.07 | 1.77 ± 0.25 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bianchi, F.; Spitaler, U.; Robatscher, P.; Vogel, R.F.; Schmidt, S.; Eisenstecken, D. Comparative Lipidomics of Different Yeast Species Associated to Drosophila suzukii. Metabolites 2020, 10, 352. https://doi.org/10.3390/metabo10090352
Bianchi F, Spitaler U, Robatscher P, Vogel RF, Schmidt S, Eisenstecken D. Comparative Lipidomics of Different Yeast Species Associated to Drosophila suzukii. Metabolites. 2020; 10(9):352. https://doi.org/10.3390/metabo10090352
Chicago/Turabian StyleBianchi, Flavia, Urban Spitaler, Peter Robatscher, Rudi F. Vogel, Silvia Schmidt, and Daniela Eisenstecken. 2020. "Comparative Lipidomics of Different Yeast Species Associated to Drosophila suzukii" Metabolites 10, no. 9: 352. https://doi.org/10.3390/metabo10090352
APA StyleBianchi, F., Spitaler, U., Robatscher, P., Vogel, R. F., Schmidt, S., & Eisenstecken, D. (2020). Comparative Lipidomics of Different Yeast Species Associated to Drosophila suzukii. Metabolites, 10(9), 352. https://doi.org/10.3390/metabo10090352