Lipidomic UPLC-MS/MS Profiles of Normal-Appearing White Matter Differentiate Primary and Secondary Progressive Multiple Sclerosis
Abstract
:1. Introduction
2. Results
2.1. Post-Mortem Case Details
2.2. Immunohistochemistry of White Matter Tissue Sections
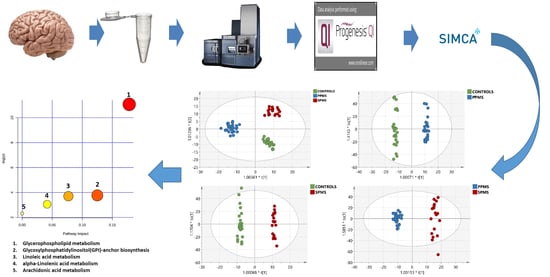
2.3. Multivariate Analysis of the RP-UPLC-TOF MSE Data
2.4. Markers of Progression between PPMS and SPMS
2.5. Lipid Markers of Progression in MS Compared with Control Cases
2.6. ROC Analysis
2.7. Metabolic Pathway Analysis
2.8. Correlation Analysis of Lipid Levels with Sex, Age, and PMI
3. Discussion
4. Materials and Methods
4.1. Post-Mortem CNS Cases
4.2. Acquisition of Lipidomic Data
4.2.1. Analytical Standards
4.2.2. CNS Sample Collection
4.2.3. Classification of WM and NAWM Tissue Using Histological and Immunohistochemical Techniques
4.2.4. Lipid Extraction from White Matter
4.2.5. UPLC–MS Data Acquisition
4.3. Data Processing and Lipid Identification
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Dilokthornsakul, P.; Valuck, R.J.; Nair, K.V.; Corboy, J.R.; Allen, R.R.; Campbell, J.D. Multiple sclerosis prevalence in the United States commercially insured population. Neurology 2016, 86, 1014–1021. [Google Scholar] [CrossRef] [Green Version]
- Confavreux, C.; Vukusic, S.; Moreau, T.; Adeleine, P. Relapses and progression of disability in multiple sclerosis. N. Engl. J. Med. 2000, 343, 1430–1438. [Google Scholar] [CrossRef] [PubMed]
- Leray, E.; Yaouanq, J.; Le Page, E.; Coustans, M.; Laplaud, D.; Oger, J.; Edan, G. Evidence for a two-stage disability progression in multiple sclerosis. Brain 2010, 133 Pt 7, 1900–1913. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scalfari, A.; Lederer, C.; Daumer, M.; Nicholas, R.; Ebers, G.C.; Muraro, P.A. The relationship of age with the clinical phenotype in multiple sclerosis. Mult. Scler. 2016, 22, 1750–1758. [Google Scholar] [CrossRef] [PubMed]
- Zeydan, B.; Kantarci, O.H. Progressive Forms of Multiple Sclerosis: Distinct Entity or Age-Dependent Phenomena. Neurol. Clin. 2018, 36, 163–171. [Google Scholar] [CrossRef] [PubMed]
- Antel, J.; Antel, S.; Caramanos, Z.; Arnold, D.L.; Kuhlmann, T. Primary progressive multiple sclerosis: Part of the MS disease spectrum or separate disease entity? Acta Neuropathol. 2012, 123, 627–638. [Google Scholar] [CrossRef] [PubMed]
- Nikic, I.; Merkler, D.; Sorbara, C.; Brinkoetter, M.; Kreutzfeldt, M.; Bareyre, F.M.; Bruck, W.; Bishop, D.; Misgeld, T.; Kerschensteiner, M. A reversible form of axon damage in experimental autoimmune encephalomyelitis and multiple sclerosis. Nat. Med. 2011, 17, 495–499. [Google Scholar] [CrossRef]
- Reynolds, R.; Roncaroli, F.; Nicholas, R.; Radotra, B.; Gveric, D.; Howell, O. The neuropathological basis of clinical progression in multiple sclerosis. Acta Neuropathol. 2011, 122, 155–170. [Google Scholar] [CrossRef]
- Choi, S.R.; Howell, O.W.; Carassiti, D.; Magliozzi, R.; Gveric, D.; Muraro, P.A.; Nicholas, R.; Roncaroli, F.; Reynolds, R. Meningeal inflammation plays a role in the pathology of primary progressive multiple sclerosis. Brain 2012, 135 Pt 10, 2925–2937. [Google Scholar] [CrossRef] [Green Version]
- Corthals, A.P. Multiple sclerosis is not a disease of the immune system. Q. Rev. Biol. 2011, 86, 287–321. [Google Scholar] [CrossRef] [Green Version]
- Neu, I.; Woelk, H. Investigations of the lipid metabolism of the white matter in multiple sclerosis: Changes in glycero-phosphatides and lipid-splitting enzymes. Neurochem. Res. 1982, 7, 727–735. [Google Scholar] [CrossRef] [PubMed]
- Budnik, V.; Ruiz-Canada, C.; Wendler, F. Extracellular vesicles round off communication in the nervous system. Nat. Rev. Neurosci. 2016, 17, 160–172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frohlich, D.; Kuo, W.P.; Fruhbeis, C.; Sun, J.J.; Zehendner, C.M.; Luhmann, H.J.; Pinto, S.; Toedling, J.; Trotter, J.; Kramer-Albers, E.M. Multifaceted effects of oligodendroglial exosomes on neurons: Impact on neuronal firing rate, signal transduction and gene regulation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2014, 369, 20130510. [Google Scholar] [CrossRef] [PubMed]
- Laule, C.; Pavlova, V.; Leung, E.; Zhao, G.; MacKay, A.L.; Kozlowski, P.; Traboulsee, A.L.; Li, D.K.; Moore, G.R. Diffusely abnormal white matter in multiple sclerosis: Further histologic studies provide evidence for a primary lipid abnormality with neurodegeneration. J. Neuropathol. Exp. Neurol. 2013, 72, 42–52. [Google Scholar] [CrossRef] [Green Version]
- Ramos, I.R.; Lyng, F.M.; Rehman, I.U.; Sharrack, B.; Woodroofe, M.N. The use of vibrational spectroscopy to study the pathogenesis multiple sclerosis and other neurological conditions. Appl. Spectrosc. Rev. 2017, 52, 868–882. [Google Scholar] [CrossRef]
- Aeinehband, S.; Brenner, P.; Stahl, S.; Bhat, M.; Fidock, M.D.; Khademi, M.; Olsson, T.; Engberg, G.; Jokinen, J.; Erhardt, S.; et al. Cerebrospinal fluid kynurenines in multiple sclerosis; relation to disease course and neurocognitive symptoms. Brain Behav. Immun. 2016, 51, 47–55. [Google Scholar] [CrossRef] [Green Version]
- Andersen, S.L.; Briggs, F.B.S.; Winnike, J.H.; Natanzon, Y.; Maichle, S.; Knagge, K.J.; Newby, L.K.; Gregory, S.G. Metabolome-based signature of disease pathology in MS. Mult. Scler. Relat. Disord. 2019, 31, 12–21. [Google Scholar] [CrossRef]
- Dickens, A.M.; Larkin, J.R.; Griffin, J.L.; Cavey, A.; Matthews, L.; Turner, M.R.; Wilcock, G.K.; Davis, B.G.; Claridge, T.D.; Palace, J.; et al. A type 2 biomarker separates relapsing-remitting from secondary progressive multiple sclerosis. Neurology 2014, 83, 1492–1499. [Google Scholar] [CrossRef] [Green Version]
- Gebregiworgis, T.; Nielsen, H.H.; Massilamany, C.; Gangaplara, A.; Reddy, J.; Illes, Z.; Powers, R. A Urinary Metabolic Signature for Multiple Sclerosis and Neuromyelitis Optica. J. Proteome Res. 2016, 15, 659–666. [Google Scholar] [CrossRef] [Green Version]
- Herman, S.; Akerfeldt, T.; Spjuth, O.; Burman, J.; Kultima, K. Biochemical Differences in Cerebrospinal Fluid between Secondary Progressive and Relapsing(-)Remitting Multiple Sclerosis. Cells 2019, 8, 84. [Google Scholar] [CrossRef] [Green Version]
- Herman, S.; Khoonsari, P.E.; Tolf, A.; Steinmetz, J.; Zetterberg, H.; Akerfeldt, T.; Jakobsson, P.J.; Larsson, A.; Spjuth, O.; Burman, J.; et al. Integration of magnetic resonance imaging and protein and metabolite CSF measurements to enable early diagnosis of secondary progressive multiple sclerosis. Theranostics 2018, 8, 4477–4490. [Google Scholar] [CrossRef] [PubMed]
- Lim, C.K.; Bilgin, A.; Lovejoy, D.B.; Tan, V.; Bustamante, S.; Taylor, B.V.; Bessede, A.; Brew, B.J.; Guillemin, G.J. Kynurenine pathway metabolomics predicts and provides mechanistic insight into multiple sclerosis progression. Sci. Rep. 2017, 7, 41473. [Google Scholar] [CrossRef] [PubMed]
- Stoessel, D.; Stellmann, J.P.; Willing, A.; Behrens, B.; Rosenkranz, S.C.; Hodecker, S.C.; Sturner, K.H.; Reinhardt, S.; Fleischer, S.; Deuschle, C.; et al. Metabolomic Profiles for Primary Progressive Multiple Sclerosis Stratification and Disease Course Monitoring. Front. Hum. Neurosci. 2018, 12, 226. [Google Scholar] [CrossRef] [PubMed]
- Cicalini, I.; Rossi, C.; Pieragostino, D.; Agnifili, L.; Mastropasqua, L.; di Ioia, M.; De Luca, G.; Onofrj, M.; Federici, L.; Del Boccio, P. Integrated Lipidomics and Metabolomics Analysis of Tears in Multiple Sclerosis: An Insight into Diagnostic Potential of Lacrimal Fluid. Int. J. Mol. Sci. 2019, 20, 1265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, G.; Hasan, M.; Kwon, O.S.; Jung, B.H. Identification of Altered Metabolic Pathways during Disease Progression in EAE Mice via Metabolomics and Lipidomics. Neuroscience 2019, 416, 74–87. [Google Scholar] [CrossRef] [PubMed]
- Lotsch, J.; Schiffmann, S.; Schmitz, K.; Brunkhorst, R.; Lerch, F.; Ferreiros, N.; Wicker, S.; Tegeder, I.; Geisslinger, G.; Ultsch, A. Machine-learning based lipid mediator serum concentration patterns allow identification of multiple sclerosis patients with high accuracy. Sci. Rep. 2018, 8, 14884. [Google Scholar] [CrossRef] [PubMed]
- Nogueras, L.; Gonzalo, H.; Jove, M.; Sol, J.; Gil-Sanchez, A.; Hervas, J.V.; Valcheva, P.; Gonzalez-Mingot, C.; Solana, M.J.; Peralta, S.; et al. Lipid profile of cerebrospinal fluid in multiple sclerosis patients: A potential tool for diagnosis. Sci. Rep. 2019, 9, 11313. [Google Scholar] [CrossRef] [Green Version]
- Oliveira, E.M.L.; Montani, D.A.; Oliveira-Silva, D.; Rodrigues-Oliveira, A.F.; Matas, S.L.A.; Fernandes, G.B.P.; Silva, I.; Lo Turco, E.G. Multiple sclerosis has a distinct lipid signature in plasma and cerebrospinal fluid. Arq. Neuropsiquiatr. 2019, 77, 696–704. [Google Scholar] [CrossRef]
- Pieragostino, D.; Cicalini, I.; Lanuti, P.; Ercolino, E.; di Ioia, M.; Zucchelli, M.; Zappacosta, R.; Miscia, S.; Marchisio, M.; Sacchetta, P.; et al. Enhanced release of acid sphingomyelinase-enriched exosomes generates a lipidomics signature in CSF of Multiple Sclerosis patients. Sci. Rep. 2018, 8, 3071. [Google Scholar] [CrossRef] [Green Version]
- Pieragostino, D.; D’Alessandro, M.; di Ioia, M.; Rossi, C.; Zucchelli, M.; Urbani, A.; Di Ilio, C.; Lugaresi, A.; Sacchetta, P.; Del Boccio, P. An integrated metabolomics approach for the research of new cerebrospinal fluid biomarkers of multiple sclerosis. Mol. Biosyst. 2015, 11, 1563–1572. [Google Scholar] [CrossRef] [Green Version]
- Podlecka-Pietowska, A.; Kacka, A.; Zakrzewska-Pniewska, B.; Nojszewska, M.; Zieminska, E.; Chalimoniuk, M.; Toczylowska, B. Altered Cerebrospinal Fluid Concentrations of Hydrophobic and Hydrophilic Compounds in Early Stages of Multiple Sclerosis-Metabolic Profile Analyses. J. Mol. Neurosci. 2019, 69, 94–105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Senanayake, V.K.; Jin, W.; Mochizuki, A.; Chitou, B.; Goodenowe, D.B. Metabolic dysfunctions in multiple sclerosis: Implications as to causation, early detection, and treatment, a case control study. BMC Neurol. 2015, 15, 154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trepanier, M.O.; Hildebrand, K.D.; Nyamoya, S.D.; Amor, S.; Bazinet, R.P.; Kipp, M. Phosphatidylcholine 36:1 concentration decreases along with demyelination in the cuprizone animal model and in post-mortem multiple sclerosis brain tissue. J. Neurochem. 2018, 145, 504–515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vergara, D.; D’Alessandro, M.; Rizzello, A.; De Riccardis, L.; Lunetti, P.; Del Boccio, P.; De Robertis, F.; Trianni, G.; Maffia, M.; Giudetti, A.M. A lipidomic approach to the study of human CD4(+) T lymphocytes in multiple sclerosis. BMC Neurosci. 2015, 16, 46. [Google Scholar] [CrossRef] [Green Version]
- Bhargava, P.; Calabresi, P.A. Metabolomics in multiple sclerosis. Mult. Scler. 2016, 22, 451–460. [Google Scholar] [CrossRef]
- Del Boccio, P.; Rossi, C.; di Ioia, M.; Cicalini, I.; Sacchetta, P.; Pieragostino, D. Integration of metabolomics and proteomics in multiple sclerosis: From biomarkers discovery to personalized medicine. Proteom. Clin. Appl. 2016, 10, 470–484. [Google Scholar] [CrossRef]
- Wheeler, D.; Bandaru, V.V.; Calabresi, P.A.; Nath, A.; Haughey, N.J. A defect of sphingolipid metabolism modifies the properties of normal appearing white matter in multiple sclerosis. Brain 2008, 131 Pt 11, 3092–3102. [Google Scholar] [CrossRef] [Green Version]
- Zhornitsky, S.; McKay, K.A.; Metz, L.M.; Teunissen, C.E.; Rangachari, M. Cholesterol and markers of cholesterol turnover in multiple sclerosis: Relationship with disease outcomes. Mult. Scler. Relat. Disord. 2016, 5, 53–65. [Google Scholar] [CrossRef]
- Bittner, S.; Ruck, T.; Schuhmann, M.K.; Herrmann, A.M.; ou Maati, H.M.; Bobak, N.; Borsotto, M. Endothelial TWIK-related potassium channel-1 (TREK1) regulates immune-cell trafficking into the CNS. Nat. Med. 2013, 19, 1161–1165. [Google Scholar] [CrossRef]
- Quehenberger, O.; Armando, A.M.; Brown, A.H.; Milne, S.B.; Myers, D.S.; Merrill, A.H.; Bandyopadhyay, S.; Jones, K.N.; Kelly, S.; Shaner, R.L.; et al. Lipidomics reveals a remarkable diversity of lipids in human plasma. J. Lipid Res. 2010, 51, 3299–3305. [Google Scholar] [CrossRef] [Green Version]
- Ferreira, H.B.; Neves, B.; Guerra, I.M.; Moreira, A.; Melo, T.; Paiva, A.; Domingues, M.R. An overview of lipidomic analysis in different human matrices of multiple sclerosis. Mult. Scler. Relat. Disord. 2020, 44, 102189. [Google Scholar] [CrossRef] [PubMed]
- Lassmann, H.; van Horssen, J.; Mahad, D. Progressive multiple sclerosis: Pathology and pathogenesis. Nat. Rev. Neurol. 2012, 8, 647–656. [Google Scholar] [CrossRef] [PubMed]
- Calabrese, M.; Favaretto, A.; Martini, V.; Gallo, P. Grey matter lesions in MS: From histology to clinical implications. Prion 2013, 7, 20–27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, K.; Baluya, D.; Tosun, M.; Li, F.; Maletic-Savatic, M. Imaging Mass Spectrometry: A New Tool to Assess Molecular Underpinnings of Neurodegeneration. Metabolites 2019, 9, 135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bergholt, M.S.; Serio, A.; McKenzie, J.S.; Boyd, A.; Soares, R.F.; Tillner, J.; Chiappini, C.; Wu, V.; Dannhorn, A.; Takats, Z.; et al. Correlated Heterospectral Lipidomics for Biomolecular Profiling of Remyelination in Multiple Sclerosis. ACS Cent Sci. 2018, 4, 39–51. [Google Scholar] [CrossRef] [Green Version]
- Rabagny, Y.; Herrmann, W.; Geisel, J.; Kirsch, S.H.; Obeid, R. Quantification of plasma phospholipids by ultra performance liquid chromatography tandem mass spectrometry. Anal. Bioanal. Chem. 2011, 401, 891–899. [Google Scholar] [CrossRef]
- Adkins, Y.; Soulika, A.M.; Mackey, B.; Kelley, D.S. Docosahexaenoic acid (22:6n-3) Ameliorated the Onset and Severity of Experimental Autoimmune Encephalomyelitis in Mice. Lipids 2019, 54, 13–23. [Google Scholar] [CrossRef] [Green Version]
- Farooqui, A.A. Lipid mediators in the neural cell nucleus: Their metabolism, signaling, and association with neurological disorders. Neuroscientist 2009, 15, 392–407. [Google Scholar] [CrossRef]
- Farooqui, A.A.; Horrocks, L.A. Phospholipase A2-generated lipid mediators in the brain: The good, the bad, and the ugly. Neuroscientist 2006, 12, 245–260. [Google Scholar] [CrossRef]
- Payne, S.G.; Oskeritzian, C.A.; Griffiths, R.; Subramanian, P.; Barbour, S.E.; Chalfant, C.E.; Milstein, S.; Spiegel, S. The immunosuppressant drug FTY720 inhibits cytosolic phospholipase A2 independently of sphingosine-1-phosphate receptors. Blood 2007, 109, 1077–1085. [Google Scholar] [CrossRef] [Green Version]
- O’Brien, J.S.; Sampson, E.L. Lipid composition of the normal human brain: Gray matter, white matter, and myelin. J. Lipid Res. 1965, 6, 537–544. [Google Scholar] [PubMed]
- Schmitt, S.; Castelvetri, L.C.; Simons, M. Metabolism and functions of lipids in myelin. Biochim. Biophys. Acta 2015, 1851, 999–1005. [Google Scholar] [CrossRef] [PubMed]
- Manzoli, F.A.; Stefoni, S.; Manzoli-Guidotti, L.; Barbieri, M. The fatty acids of myelin phospholipids. FEBS Lett. 1970, 10, 317–320. [Google Scholar] [CrossRef] [Green Version]
- Rioux, F.M.; Innis, S.M. Oleic acid (18:1) in plasma, liver and brain myelin lipid of piglets fed from birth with formulas differing in 18:1 content. J. Nutr. 1992, 122, 1521–1528. [Google Scholar] [CrossRef] [PubMed]
- Baker, R.W.; Thompson, R.H.; Zilkha, K.J. Fatty-acid composition of brain lecithins in multiple sclerosis. Lancet 1963, 1, 26–27. [Google Scholar] [CrossRef]
- McNamara, R.K.; Rider, T.; Jandacek, R.; Tso, P. Abnormal fatty acid pattern in the superior temporal gyrus distinguishes bipolar disorder from major depression and schizophrenia and resembles multiple sclerosis. Psychiatry Res. 2014, 215, 560–567. [Google Scholar] [CrossRef] [Green Version]
- Timmers, S.; de Vogel-van den Bosch, J.; Hesselink, M.K.; van Beurden, D.; Schaart, G.; Ferraz, M.J.; Losen, M.; Martinez-Martinez, P.; De Baets, M.H.; Aerts, J.M.; et al. Paradoxical increase in TAG and DAG content parallel the insulin sensitizing effect of unilateral DGAT1 overexpression in rat skeletal muscle. PLoS ONE 2011, 6, e14503. [Google Scholar] [CrossRef] [Green Version]
- Penesova, A.; Vlcek, M.; Imrich, R.; Vernerova, L.; Marko, A.; Meskova, M.; Grunnerova, L.; Turcani, P.; Jezova, D.; Kollar, B. Hyperinsulinemia in newly diagnosed patients with multiple sclerosis. Metab. Brain Dis. 2015, 30, 895–901. [Google Scholar] [CrossRef]
- Nakajima, S.; Gotoh, M.; Fukasawa, K.; Murakami-Murofushi, K.; Kunugi, H. Oleic acid is a potent inducer for lipid droplet accumulation through its esterification to glycerol by diacylglycerol acyltransferase in primary cortical astrocytes. Brain Res. 2019, 1725, 146484. [Google Scholar] [CrossRef]
- Walter, S.; Fassbender, K. Sphingolipids in Multiple Sclerosis. Cell. Physiol. Biochem. 2010, 26, 49–56. [Google Scholar] [CrossRef]
- Dargahi, N.; Katsara, M.; Tselios, T.; Androutsou, M.E.; de Courten, M.; Matsoukas, J.; Apostolopoulos, V. Multiple Sclerosis: Immunopathology and Treatment Update. Brain Sci. 2017, 7, 78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vidaurre, O.G.; Haines, J.D.; Katz Sand, I.; Adula, K.P.; Huynh, J.L.; McGraw, C.A.; Zhang, F.; Varghese, M.; Sotirchos, E.; Bhargava, P.; et al. Cerebrospinal fluid ceramides from patients with multiple sclerosis impair neuronal bioenergetics. Brain 2014, 137 Pt 8, 2271–2286. [Google Scholar] [CrossRef] [PubMed]
- Moscatelli, E.A.; Isaacson, E. Gas liquid chromatographic analysis of sphingosine bases in sphingolipids of human normal and multiple sclerosis cerebral white matter. Lipids 1969, 4, 550–555. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, B.; West, J.A.; Koulman, A. A review of odd-chain fatty acid metabolism and the role of pentadecanoic Acid (c15:0) and heptadecanoic Acid (c17:0) in health and disease. Molecules 2015, 20, 2425–2444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pfeuffer, M.; Jaudszus, A. Pentadecanoic and Heptadecanoic Acids: Multifaceted Odd-Chain Fatty Acids. Adv. Nutr. 2016, 7, 730–734. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waller, R.; Woodroofe, M.N.; Wharton, S.B.; Ince, P.G.; Francese, S.; Heath, P.R.; Cudzich-Madry, A.; Thomas, R.H.; Rounding, N.; Sharrack, B.; et al. Gene expression profiling of the astrocyte transcriptome in multiple sclerosis normal appearing white matter reveals a neuroprotective role. J. Neuroimmunol. 2016, 299, 139–146. [Google Scholar] [CrossRef]
- Astarita, G.; Stocchero, M.; Paglia, G. Unbiased Lipidomics and Metabolomics of Human Brain Samples. Methods Mol. Biol. 2018, 1750, 255–269. [Google Scholar]
- Vorkas, P.A. Expanding lipidome coverage using MS/MS-aided untargeted data-independent RP-UPLC-TOF-MS(E) acquisition. Bioanalysis 2018, 10, 307–319. [Google Scholar] [CrossRef] [Green Version]
- Plumb, R.S.; Johnson, K.A.; Rainville, P.; Smith, B.W.; Wilson, I.D.; Castro-Perez, J.M.; Nicholson, J.K. UPLC/MS(E); a new approach for generating molecular fragment information for biomarker structure elucidation. Rapid Commun. Mass Spectrom. 2006, 20, 1989–1994. [Google Scholar] [CrossRef]
- Gika, H.G.; Theodoridis, G.A.; Earll, M.; Wilson, I.D. A QC approach to the determination of day-to-day reproducibility and robustness of LC-MS methods for global metabolite profiling in metabonomics/metabolomics. Bioanalysis 2012, 4, 2239–2247. [Google Scholar] [CrossRef]
- Gika, H.G.; Zisi, C.; Theodoridis, G.; Wilson, I.D. Protocol for quality control in metabolic profiling of biological fluids by U(H)PLC-MS. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2016, 1008, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Theodoridis, G.; Gika, H.G.; Wilson, I.D. Mass spectrometry-based holistic analytical approaches for metabolite profiling in systems biology studies. Mass Spectrom. Rev. 2011, 30, 884–906. [Google Scholar] [CrossRef] [PubMed]
- Kirwan, G.M.; Johansson, E.; Kleemann, R.; Verheij, E.R.; Wheelock, A.M.; Goto, S.; Trygg, J.; Wheelock, C.E. Building multivariate systems biology models. Anal. Chem. 2012, 84, 7064–7071. [Google Scholar] [CrossRef] [PubMed]
- Trygg, J.; Holmes, E.; Lundstedt, T. Chemometrics in metabonomics. J. Proteome Res. 2007, 6, 469–479. [Google Scholar] [CrossRef] [PubMed]
- Wheelock, A.M.; Wheelock, C.E. Trials and tribulations of ’omics data analysis: Assessing quality of SIMCA-based multivariate models using examples from pulmonary medicine. Mol. Biosyst. 2013, 9, 2589–2596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, Z.; Chen, E.; Ouyang, X.; Xu, X.; Ma, S.; Ji, F.; Wu, D.; Zhang, S.; Zhao, Y.; Li, L. Metabolomics and Cytokine Analysis for Identification of Severe Drug-Induced Liver Injury. J. Proteome Res. 2019, 18, 2514–2524. [Google Scholar] [CrossRef]
- Smith, C.A.; O’Maille, G.; Want, E.J.; Qin, C.; Trauger, S.A.; Brandon, T.R.; Custodio, D.E.; Abagyan, R.; Siuzdak, G. METLIN: A metabolite mass spectral database. Drug Monit. 2005, 27, 747–751. [Google Scholar] [CrossRef]
- Wishart, D.S.; Knox, C.; Guo, A.C.; Eisner, R.; Young, N.; Gautam, B.; Hau, D.D.; Psychogios, N.; Dong, E.; Bouatra, S.; et al. HMDB: A knowledgebase for the human metabolome. Nucleic Acids Res. 2009, 37 (Suppl. S1), D603–D610. [Google Scholar] [CrossRef]
- Fahy, E.; Sud, M.; Cotter, D.; Subramaniam, S. LIPID MAPS online tools for lipid research. Nucleic Acids Res. 2007, 35, W606–W612. [Google Scholar] [CrossRef] [Green Version]
- Cajka, T.; Fiehn, O. LC-MS-Based Lipidomics and Automated Identification of Lipids Using the LipidBlast In-Silico MS/MS Library. Methods Mol. Biol. 2017, 1609, 149–170. [Google Scholar]
- Kind, T.; Liu, K.H.; Lee, D.Y.; DeFelice, B.; Meissen, J.K.; Fiehn, O. LipidBlast in silico tandem mass spectrometry database for lipid identification. Nat. Methods 2013, 10, 755–758. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chong, J.; Soufan, O.; Li, C.; Caraus, I.; Li, S.; Bourque, G.; Wishart, D.S.; Xia, J. MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018, 46, W486–W494. [Google Scholar] [CrossRef] [PubMed] [Green Version]







| Healthy Controls (n = 8) | PPMS (n = 9) | SPMS (n = 7) | p-Value | |
|---|---|---|---|---|
| Age (years) | 76 ± 9 | 61 ± 10 | 60 ± 15 | n.s. |
| Range | (63–88) | (39–73) | (39–88) | |
| Female/Male | 2/6 | 6/3 | 6/1 | n.s. |
| PMI (hours) | 22.3 ± 8.5 | 14.6 ± 5.1 | 13.7 ± 6.3 | n.s. |
| (5–33) | (8–24) | (7–24) |
| Peak Number | MS Mode | RT | m/z | Adduct | Lipid Identified | Delta (ppm) | p-Value | FC | Trend (SPMS vs. PPMS) | RSD(%) in QCs |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | pos | 14.38 | 778.6076 | M + Na | CerP (d18: 1/26: 1) | −1.123 | 0.04439 | 1.210 | ↑ | 21.65 |
| 2 | pos | 6.93 | 621.4856 | M + K | DG (18: 0_15: 0) | 0.176 | 0.00954 | 1.338 | ↑ | 11.19 |
| 3 | pos | 14.8 | 635.5615 | M + H | DG (18: 2_19: 0) | 0.970 | 0.02617 | 1.387 | ↑ | 27.39 |
| 4 | pos | 0.61 | 429.2722 | M + NH4 | lysoPC (10: 0) | −0.506 | 0.01166 | 2.144 | ↑ | 27.30 |
| 5 | pos | 1.84 | 527.3813 | M + NH4 | lysoPC (17: 0) | −1.301 | 0.03450 | 0.775 | ↓ | 18.24 |
| 6 | neg | 6.21 | 514.3291 | M − H2O − H | lysoPE (22: 2) | −2.182 | 0.01040 | 1.521 | ↑ | 24.22 |
| 7 | neg | 13.73 | 683.5004 | M − H2O − H | PA (18: 1_18: 0) | −2.372 | 0.02982 | 1.182 | ↑ | 17.39 |
| 8 | neg | 10.15 | 779.5376 | M + Cl | PA (18: 1_21: 0) | 1.749 | 0.00914 | 0.750 | ↓ | 27.26 |
| 9 | neg | 13.73 | 733.5153 | M − H2O − H | PA (20: 4_20: 0) | −3.302 | 0.02392 | 1.418 | ↑ | 27.47 |
| 10 | neg | 6.77 | 885.6538 | M + FA − H | PA (22: 2_24: 0) | −6.192 | 0.02793 | 0.709 | ↓ | 17.69 |
| 11 | pos | 9.79 | 746.4815n | M + H, M + NH4 | PA (22: 6_18: 1) | −9.618 | 0.00450 | 0.762 | ↓ | 18.72 |
| 12 | pos | 8.43 | 821.5770 | M + NH4 | PC (20: 5_18: 2) | −4.189 | 0.03984 | 1.497 | ↑ | 19.01 |
| 13 | pos | 13.66 | 794.6051 | M + Na | PC (P-18: 0_18: 1) | 2.244 | 0.01342 | 0.747 | ↓ | 12.38 |
| 14 | neg | 13.66 | 780.593 | M − H2O − H | PE (18: 1_22: 1) | 2.113 | 0.02526 | 0.745 | ↓ | 25.62 |
| 15 | neg | 7.64 | 696.5034 | M − H2O − H | PE (18: 2_16: 0) | 8.412 | 0.00112 | 0.587 | ↓ | 21.90 |
| 16 | neg | 11.22 | 820.5576 | M + Cl | PE (18: 2_21: 0) | −6.698 | 0.01061 | 0.796 | ↓ | 13.39 |
| 17 | neg | 8.92 | 832.564 | M + Cl | PE (18: 2_22: 1) | 1.436 | 0.01213 | 0.793 | ↓ | 26.52 |
| 18 | neg | 13.51 | 766.5446 | M − H | PE (20: 2_18: 2) | 7.043 | 0.00175 | 0.702 | ↓ | 17.09 |
| 19 | neg | 13.57 | 792.5594 | M − H | PE (20: 3_20: 2) | 5.652 | 0.01380 | 0.756 | ↓ | 27.45 |
| 20 | neg | 13.93 | 806.6094 | M − H2O − H | PE (20: 3_22: 0) | 3.015 | 0.01864 | 0.745 | ↓ | 18.16 |
| 21 | neg | 8.49 | 812.5500 | M + FA − H | PE (20: 4_18: 0) | 6.873 | 0.00507 | 0.515 | ↓ | 18.51 |
| 22 | neg | 10.28 | 840.5818 | M + FA − H | PE (20: 4_20: 0) | 7.300 | 0.02022 | 0.660 | ↓ | 11.71 |
| 23 | neg | 7.71 | 702.5488 | M − H | PE (P-16: 0_18: 0) | 6.340 | 0.01840 | 0.772 | ↓ | 18.57 |
| 24 | neg | 7.57 | 696.4689 | M + Cl | PE (P-18: 0_13: 0) | −7.776 | 0.00027 | 0.528 | ↓ | 28.00 |
| 25 | neg | 7.78 | 762.5722 | M + FA − H | PE (P-18: 0_17: 0) | 9.407 | 0.01114 | 0.764 | ↓ | 13.19 |
| 26 | neg | 13.51 | 796.5545 | M + FA − H | PE (P-18: 0_20: 4) | 6.307 | 0.02060 | 1.223 | ↑ | 27.33 |
| 27 | neg | 10.93 | 794.5401 | M + FA − H | PE (P-18: 0_20: 5) | 7.929 | 0.00260 | 1.300 | ↑ | 16.75 |
| 28 | neg | 11.58 | 820.597 | M + Cl | PE (P-18: 0_22: 1) | −2.904 | 0.00037 | 0.723 | ↓ | 18.92 |
| 29 | neg | 10.44 | 802.5798 | M − H | PE (P-20: 0_22: 6) | 5.269 | 0.00039 | 0.663 | ↓ | 9.27 |
| 30 | neg | 10.51 | 785.5129 | M + Cl | PG (18: 0_16: 0) | 3.202 | 0.00092 | 1.751 | ↑ | 10.77 |
| 31 | neg | 10.93 | 811.5285 | M + Cl | PG (18: 1_18: 0) | 3.008 | 0.00076 | 1.485 | ↑ | 16.55 |
| 32 | neg | 6.21 | 847.5457 | M − H | PG (22: 6_20: 1) | −4.431 | 0.00073 | 0.573 | ↓ | 21.41 |
| 33 | neg | 4.35 | 905.5312 | M + FA − H | PI (18: 2_18: 1) | −9.876 | 0.03147 | 0.751 | ↓ | 10.79 |
| 34 | neg | 12.43 | 889.5749 | M − H | PI (22: 2_16: 0) | −7.040 | 0.00121 | 0.702 | ↓ | 11.41 |
| 35 | neg | 10.08 | 878.6138 | M + FA − H | PS (18: 0_21: 0) | 1.210 | 0.01828 | 0.700 | ↓ | 16.67 |
| 36 | neg | 8.56 | 792.5255 | M − H2O − H | PS (18: 1_20: 3) | 8.655 | 0.00255 | 1.626 | ↑ | 19.32 |
| 37 | neg | 9.28 | 826.5966 | M − H2O − H | PS (18: 1_22: 0) | −0.113 | 0.00101 | 0.701 | ↓ | 14.02 |
| 38 | neg | 9.14 | 916.6283 | M + FA − H | PS (18: 1_24: 1) | −0.160 | 0.00014 | 0.546 | ↓ | 16.38 |
| 39 | neg | 7.78 | 876.5505 | M + Cl | PS (18: 2_22: 1) | −2.583 | 0.00258 | 0.641 | ↓ | 19.13 |
| 40 | neg | 7.84 | 816.5792 | M − H | PS (20: 1_18: 0) | 3.884 | 0.04284 | 0.717 | ↓ | 14.43 |
| 41 | neg | 9.28 | 890.5644 | M + Cl | PS (20: 3_21: 0) | −4.546 | 0.00035 | 0.647 | ↓ | 22.84 |
| 42 | neg | 11 | 862.4866 | M + FA − H | PS (22: 6_17: 2) | −1.232 | 0.00484 | 1.390 | ↑ | 24.58 |
| 43 | neg | 12.14 | 880.5401 | M + FA − H | PS (22: 6_18: 0) | 6.702 | 0.04395 | 0.844 | ↓ | 18.81 |
| 44 | neg | 10.93 | 811.4895 | M + FA − H | SQDG (18: 0_12: 0) | 1.591 | 0.02255 | 1.306 | ↑ | 26.20 |
| p Value | FC | Trend | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Peak Number | MS Mode | RT | m/z | Adduct | Lipid Identified | Delta (ppm) | CON a-PPMS | CON-SPMS | PPMS/CON | SPMS/CON | PPMS/CON | SPMS/CON | RSD% in QCs |
| 1 | pos | 3.05 | 719.5589 | M + Na | DG (20: 4_22: 2) | 0.526 | 0.0448 | 0.0238 | 0.875 | 0.853 | ↓ | ↓ | 15.86 |
| 2 | pos | 8.36 | 704.5254 | M + NH4 | PA (18: 2_17: 0) | 4.296 | 0.0096 | 0.0025 | 0.796 | 0.731 | ↓ | ↓ | 15.57 |
| 3 | pos | 6.28 | 738.5091 | M + NH4 | PA (20: 5_18: 1) | 3.127 | 0.0376 | 0.0102 | 0.585 | 0.472 | ↓ | ↓ | 17.74 |
| 4 | neg | 12.14 | 752.5621 | M − H2O − H | PE (18: 2_20: 0) | 2.771 | 0.0003 | 0.0065 | 1.553 | 1.540 | ↑ | ↑ | 25.34 |
| 5 | neg | 13.86 | 840.5768 | M + FA − H | PE (20: 4_20: 0) | 0.943 | 0.0017 | 0.0020 | 1.306 | 1.315 | ↑ | ↑ | 17.26 |
| 6 | neg | 8.72 | 828.5276 | M + Cl | PE (20: 4_20: 1) | −5.046 | 0.0105 | 0.0053 | 0.589 | 0.549 | ↓ | ↓ | 25.55 |
| 7 | neg | 10.71 | 882.5863 | M + Cl | PE (22: 6_22:0) | 9.208 | 0.0444 | 0.0164 | 0.671 | 0.588 | ↓ | ↓ | 24.42 |
| 8 | neg | 8.2 | 819.5428 | M − H2O − H | PI (18:0_16:0) | 4.231 | 0.0163 | 0.0079 | 0.621 | 0.583 | ↓ | ↓ | 21.29 |
| 9 | neg | 8.36 | 846.5546 | M + FA − H | PS (18:2_19:0) | 5.461 | 0.0102 | 0.0092 | 0.786 | 0.799 | ↓ | ↓ | 15.08 |
| 10 | neg | 8.85 | 880.5744 | M + FA − H | Sulfatide (d18: 1/20: 0) | −9.753 | 0.0111 | 0.0040 | 0.759 | 0.729 | ↓ | ↓ | 20.69 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pousinis, P.; Ramos, I.R.; Woodroofe, M.N.; Cole, L.M. Lipidomic UPLC-MS/MS Profiles of Normal-Appearing White Matter Differentiate Primary and Secondary Progressive Multiple Sclerosis. Metabolites 2020, 10, 366. https://doi.org/10.3390/metabo10090366
Pousinis P, Ramos IR, Woodroofe MN, Cole LM. Lipidomic UPLC-MS/MS Profiles of Normal-Appearing White Matter Differentiate Primary and Secondary Progressive Multiple Sclerosis. Metabolites. 2020; 10(9):366. https://doi.org/10.3390/metabo10090366
Chicago/Turabian StylePousinis, Petros, Ines R. Ramos, M. Nicola Woodroofe, and Laura M. Cole. 2020. "Lipidomic UPLC-MS/MS Profiles of Normal-Appearing White Matter Differentiate Primary and Secondary Progressive Multiple Sclerosis" Metabolites 10, no. 9: 366. https://doi.org/10.3390/metabo10090366
APA StylePousinis, P., Ramos, I. R., Woodroofe, M. N., & Cole, L. M. (2020). Lipidomic UPLC-MS/MS Profiles of Normal-Appearing White Matter Differentiate Primary and Secondary Progressive Multiple Sclerosis. Metabolites, 10(9), 366. https://doi.org/10.3390/metabo10090366