Untargeted Metabolomics Studies on Drug-Incubated Phragmites australis Profiles
Abstract
:1. Introduction
2. Results
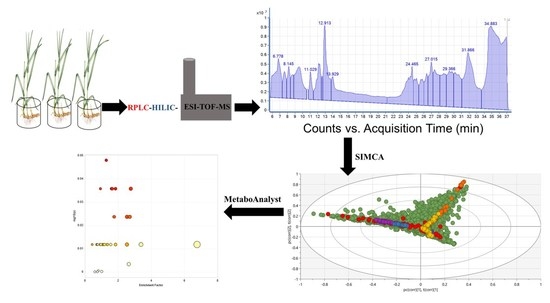
2.1. Metabolic Profiling Elucidation in Phragmites australis Extracts with RPLC-HILIC-ESI-TOF-MS
2.2. Different Extracts of Phragmites australis’s Metabolic Fingerprints
2.3. Phragmites australis Leaf, Rhizome and Root Metabolic Fingerprints
2.4. Untargeted Metabolomics Analysis of Phragmites australis Incubated with DCF or CBZ
2.5. Metabolism of Diclofenac in Phragmites australis
2.6. Metabolism of Carbamazepine in Phragmites australis
2.7. Impacts of DCF and CBZ on Phragmites australis Metabolic Pathways
3. Discussion
4. Materials and Methods
4.1. Reagents and Chemicals
4.2. Plant Samples
4.3. Extraction
4.4. Instruments
4.5. Quality Control of the RPLC-HILIC-ESI-TOF-MS System
4.6. Data Evaluation
4.6.1. Spectrometric Data Evaluation
4.6.2. DCF and CBZ Transformation Products Detection
4.6.3. Statistical Data Analysis
- Metabolite fingerprinting was used to capture metabolite patterns across metabolite profiles. They are characterized without further identification steps (i.e., without need for standard reference material). Partial Least Squares (PLS) and Orthogonal Partial Least Squares regression-Discriminant Analysis (OPLS-DA) were used to relate sets of X-variables (such as plant part, plant number, extraction solvent and drug incubation) to the metabolites matrix. SIMCA 16 has a tool called Multiblock Orthogonal Component Analysis (MOCA). MOCA’s concept is used to accomplish a fast and accurate analysis of multiple blocks of data (variables) registered for the same set of observations. MOCA aims at extracting the information in complex multi-block data analytics. Furthermore, it will extract two sets of components: the joint and the unique components. The quality of the models is described by R2 and Q2 values, where R2 is the proportion of variance in the data explained by the models and indicates the goodness of fit and Q2 is the proportion of variance in the data predictable by the model and expresses predictability [41].
- Metabolite profiling which uses sets of predefined metabolites were studied in different samples of P. australis and differences in metabolites were usually related to the incubation with DCF or CBZ. Metabolite/variable selection was conducted to observe only the most significant metabolite candidates that explain the differences between the samples using S- and contribution-plots. The statistical models were built with confidence limits at 95%. Also, the differentiating metabolic profile (DMF) was chosen based on their contribution to the variation and correlation within the data sets. The related metabolic pathways were analyzed using MetaboAnalyst 4.0. Moreover, their contributions and biological clarifications were described based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. The KEGG pathway analysis tool was used by the Arabidopsis thaliana database. The pathway analysis module combines the enrichment analysis and topology analysis based on KEGG. Fisher’s test was used to generate p values. The p value was equal to 0.05, which indicates the fundamental connection of the identified metabolite with their respective metabolite and not due to the random chance [42,43].
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tohge, T.; Nishiyama, Y.; Hirai, M.Y.; Yano, M.; Nakajima, J.-I.; Awazuhara, M.; Inoue, E.; Takahashi, H.; Goodenowe, D.B.; Kitayama, M.; et al. Identification of Genes Involved in Anthocyanin Accumulation by Integrated Analysis of Metabolome and Transcriptome in Pap1-Overexpressing Arabidopsis Plants; Springer: Dordrecht, The Netherlands, 2007; pp. 159–168. [Google Scholar]
- Sandermann, H.; Diesperger, H.; Scheel, D. Metabolism of Xenobiotics by Plant Cell Cultures; Springer: Berlin/Heidelberg, Germany, 1977; pp. 178–196. [Google Scholar]
- Schröder, P.; Scheer, P.; Belford, E.J.D. Metabolism of organic xenobiotics in plants: Conjugating enzymes and metabolic endpoints. Minerva Biotecnol. 2002, 13, 85–91. [Google Scholar]
- Schröder, P.; Maier, H.; Debus, R. Detoxification of Herbicides in Phragmites australis. Z. Nat. C 2005, 60, 317. [Google Scholar] [CrossRef]
- Villette, C.; Maurer, L.; Wanko, A.; Heintz, D. Xenobiotics metabolization in Salix alba leaves uncovered by mass spectrometry imaging. Metabolomics 2019, 15, 122. [Google Scholar] [CrossRef]
- Letzel, M.; Metzner, G.; Letzel, T. Exposure assessment of the pharmaceutical diclofenac based on long-term measurements of the aquatic input. Environ. Int. 2009, 35, 363–368. [Google Scholar] [CrossRef] [PubMed]
- Sauvêtre, A.; Schröder, P. Uptake of carbamazepine by rhizomes and endophytic bacteria of Phragmites australis. Front. Plant Sci. 2015, 6, 83. [Google Scholar] [CrossRef] [PubMed]
- Thelusmond, J.-R.; Kawka, E.; Strathmann, T.J.; Cupples, A.M. Diclofenac, carbamazepine and triclocarban biodegradation in agricultural soils and the microorganisms and metabolic pathways affected. Sci. Total Environ. 2018, 640–641, 1393–1410. [Google Scholar] [CrossRef] [PubMed]
- Vieno, N.; Sillanpää, M. Fate of diclofenac in municipal wastewater treatment plant—A review. Environ. Int. 2014, 69, 28–39. [Google Scholar] [CrossRef] [PubMed]
- Hai, F.I.; Yang, S.; Asif, M.B.; Sencadas, V.; Shawkat, S.; Sanderson-Smith, M.; Gorman, J.; Xu, Z.-Q.; Yamamoto, K. Carbamazepine as a Possible Anthropogenic Marker in Water: Occurrences, Toxicological Effects, Regulations and Removal by Wastewater Treatment Technologies. Water 2018, 10, 107. [Google Scholar] [CrossRef] [Green Version]
- Huber, C.; Bartha, B.; Schröder, P. Metabolism of diclofenac in plants—hydroxylation is followed by glucose conjugation. J. Hazard. Mater. 2012, 243, 250–256. [Google Scholar] [CrossRef]
- Park, M.G.; Blossey, B. Importance of plant traits, and herbivory for invasiveness of Phragmites australis (Poaceae). Am. J. Bot. 2008, 95, 1557–1568. [Google Scholar] [CrossRef]
- Gray, K.R.; Biddlestone, A.J. Engineered reed-bed systems for wastewater treatment. Trends Biotechnol. 1995, 13, 248–252. [Google Scholar] [CrossRef]
- Zhou, B.; Xiao, J.F.; Tuli, L.; Ressom, H.W. LC-MS-based metabolomics. Mol. Biosyst. 2012, 8, 470–481. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beale, D.; Pinu, F.; Kouremenos, K.; Poojary, M.; Narayana, V.; Boughton, B.; Kanojia, K.; Dayalan, S.; Jones, O.; Dias, D. Review of recent developments in GC–MS approaches to metabolomics-based research. Metabolomics 2018, 14, 152. [Google Scholar] [CrossRef]
- Emwas, A.-H.; Roy, R.; McKay, R.T.; Tenori, L.; Saccenti, E.; Gowda, G.A.N.; Raftery, D.; Alahmari, F.; Jaremko, L.; Jaremko, M.; et al. NMR Spectroscopy for Metabolomics Research. Metabolites 2019, 9, 123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aretz, I.; Meierhofer, D. Advantages, and pitfalls of mass spectrometry-based metabolome profiling in systems biology. Int. J. Mol. Sci. 2016, 17, 632. [Google Scholar] [CrossRef] [Green Version]
- Wahman, R.; Grassmann, J.; Schröder, P.; Letzel, T. Plant metabolomic workflows using reversed-phase LC and HILIC with ESI-TOF-MS. LCGC N. Am. 2019, 37, 8–15. [Google Scholar]
- Riach, A.C.; Perera, M.V.L.; Florance, H.V.; Penfield, S.D.; Hill, J.K. Analysis of plant leaf metabolites reveals no common response to insect herbivory by Pieris rapae in three related host-plant species. J. Exp. Bot. 2015, 66, 2547–2556. [Google Scholar] [CrossRef] [Green Version]
- Gromski, P.S.; Xu, Y.; Kotze, H.L.; Correa, E.; Ellis, D.I.; Armitage, E.G.; Turner, M.L.; Goodacre, R. Influence of missing values substitutes on multivariate analysis of metabolomics data. Metabolites 2014, 4, 433–452. [Google Scholar] [CrossRef] [Green Version]
- Lamichhane, S.; Sen, P.; Dickens, A.M.; Hyötyläinen, T.; Orešič, M. Chapter Fourteen—An Overview of metabolomics data analysis: Current tools and future perspectives. Compr. Anal. Chem. 2018, 82, 387–413. [Google Scholar]
- Prinsloo, G.; Vervoort, J. Identifying anti-HSV compounds from unrelated plants using NMR and LC-MS metabolomic analysis. Metabolomics 2018, 14, 134. [Google Scholar] [CrossRef]
- Worley, B.; Powers, R. Multivariate Analysis in Metabolomics. Curr. Metab. 2013, 1, 92–107. [Google Scholar]
- Bieber, S.; Greco, G.; Grosse, S.; Letzel, T. RPLC-HILIC, and SFC with mass spectrometry: Polarity-extended organic molecule screening in environmental (Water) samples. Anal. Chem. 2017, 89, 7907–7914. [Google Scholar] [CrossRef] [PubMed]
- Greco, G.; Grosse, S.; Letzel, T. Serial coupling of reversed-phase and zwitterionic hydrophilic interaction LC/MS for the analysis of polar and nonpolar phenols in wine. J. Sep. Sci. 2013, 36, 1379–1388. [Google Scholar] [CrossRef] [PubMed]
- Wahman, R.; Graßmann, J.; Sauvêtre, A.; Schröder, P.; Letzel, T. Lemna minor studies under various storage periods using extended-polarity extraction and metabolite non-target screening analysis. J. Pharm. Biomed. Anal. 2020, 188, 113362. [Google Scholar] [CrossRef]
- Rajab, M.; Greco, G.; Heim, C.; Helmreich, B.; Letzel, T. Serial coupling of RP and zwitterionic hydrophilic interaction LC-MS: Suspects screening of diclofenac transformation products by oxidation with a boron-doped diamond electrode. J. Sep. Sci. 2013, 36, 3011–3018. [Google Scholar] [CrossRef]
- Sauvetre, A.; May, R.; Harpaintner, R.; Poschenrieder, C.; Schröder, P. Metabolism of carbamazepine in plant roots and endophytic rhizobacteria isolated from Phragmites australis. J. Hazard. Mater. 2018, 342, 85–95. [Google Scholar] [CrossRef] [Green Version]
- Hess, D. Plant Physiology; Springer: Berlin/Heidelberg, Germany, 1975; p. 334. [Google Scholar]
- Riemenschneider, C.; Seiwert, B.; Schwarz, D.; Reemtsma, T. Extensive Transformation of the pharmaceutical carbamazepine following uptake into intact tomato plants. Environ. Sci. Technol. 2017, 51, 6100–6109. [Google Scholar] [CrossRef]
- Riemenschneider, C.; Seiwert, B.; Goldstein, M.; Al-Raggad, M.; Salameh, E.; Chefetz, B.; Reemtsma, T. An LC-MS/MS method for the determination of 28 polar environmental contaminants and metabolites in vegetables irrigated with treated municipal wastewater. Anal. Methods 2017, 9, 1273–1281. [Google Scholar] [CrossRef]
- Martínez-Piernas, A.B.; Nahim-Granados, S.; Polo-López, M.I.; Fernández-Ibáñez, P.; Murgolo, S.; Mascolo, G.; Agüera, A. Identification of transformation products of carbamazepine in lettuce crops irrigated with Ultraviolet-C treated water. Environ. Pollut. 2019, 247, 1009–1019. [Google Scholar] [CrossRef]
- Tybring, G.; von Bahr, C.; Bertilsson, L.; Collste, H.; Glaumann, H.; Solbrand, M. Metabolism of carbamazepine and its epoxide metabolite in human and rat liver in vitro. Drug Metab. Dispos. 1981, 9, 561–564. [Google Scholar]
- Lee, D.-K.; Ahn, S.; Cho, H.Y.; Yun, H.Y.; Park, J.H.; Lim, J.; Lee, J.; Kwon, S.W. Metabolic response induced by parasitic plant-fungus interactions hinder amino sugar and nucleotide sugar metabolism in the host. Sci. Rep. 2016, 6, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hurtado, C.; Parastar, H.; Matamoros, V.; Piña, B.; Tauler, R.; Bayona, J.M. Linking the morphological and metabolomic response of Lactuca sativa L exposed to emerging contaminants using GC × GC-MS and chemometric tools. Sci. Rep. 2017, 7, 6546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tantikanjana, T.; Mikkelsen, M.D.; Hussain, M.; Halkier, B.A.; Sundaresan, V. Functional Analysis of the tandem-duplicated P450 genes SPS/BUS/CYP79F1 and CYP79F2 in glucosinolate biosynthesis and plant development by Ds transposition-generated double mutants. Plant Physiol. 2004, 135, 840–848. [Google Scholar] [CrossRef] [Green Version]
- Zrenner, R.; Stitt, M.; Sonnewald, U.; Boldt, R. Pyrimidine and purine biosynthesis and degradation in plants. Annu. Rev. Plant Biol. 2006, 57, 805–836. [Google Scholar] [CrossRef] [PubMed]
- Gong, B.; Sun, S.; Yan, Y.; Jing, X.; Shi, Q. Glutathione Metabolism and Its Function in Higher Plants Adapting to Stress. In Antioxidants and Antioxidant Enzymes in Higher Plants; Springer International Publishing: Heidlberg, Germany, 2018; pp. 181–205. [Google Scholar]
- Sivaram, A.K.; Subashchandrabose, S.R.; Logeshwaran, P.; Lockington, R.; Naidu, R.; Megharaj, M. Metabolomics reveals defensive mechanisms adapted by maize on exposure to high molecular weight polycyclic aromatic hydrocarbons. Chemosphere 2019, 214, 771–780. [Google Scholar] [CrossRef] [PubMed]
- Araújo, W.L.; Nunes-Nesi, A.; Nikoloski, Z.; Sweetlove, L.J.; Fernie, A.R. Metabolic control and regulation of the tricarboxylic acid cycle in photosynthetic and heterotrophic plant tissues. Plant Cell Environ. 2012, 35, 1–21. [Google Scholar] [CrossRef] [Green Version]
- Löfstedt, T.; Trygg, J. OnPLS—A novel multiblock method for the modeling of predictive and orthogonal variation. J. Chemom. 2011, 25, 441–455. [Google Scholar]
- Chong, J.; Wishart, D.S.; Xia, J. Using MetaboAnalyst 4.0 for Comprehensive and Integrative Metabolomics Data Analysis. Curr. Protoc. Bioinform. 2019, 68, e86. [Google Scholar] [CrossRef]
- Chong, J.; Yamamoto, M.; Xia, J. MetaboAnalystR 2.0: From Raw Spectra to Biological Insights. Metabolites 2019, 9, 57. [Google Scholar] [CrossRef] [Green Version]









| DCF Transformed Products | Name | Mono Isotopic Mass (Da) (L) (Rajab, Greco et al. 2013) | Mean Mono Isotopic Mass (Da) (Ph) | Δppm | Mean RT (Min) (Ph) | SD of RT (Min) | RSD | LogD (pH = 7.4) | Leaf | Rhizome | Root |
|---|---|---|---|---|---|---|---|---|---|---|---|
| DM_1 | 2-Hydroxypropanoic acid | 152.0473 | 152.0475 | −0.99 | 8.1 | 0.05 | 0.67 | −1.86 | √ | √ | √ |
| DM_2 | 2-(Hydroxymethyl)benzene-1,4-diol | 140.0473 | 140.0472 | 0.71 | 12.5 | 0.06 | 0.51 | 0.60 | √ | √ | √ |
| DM_3 | 2-Hydroxysuccinic acid | 134.0215 | 134.0215 | 0.07 | 12.6 | 0.03 | 0.23 | −6.81 | √ | √ | √ |
| DM_4 | Succinic acid | 118.0266 | 118.0271 | −3.95 | 6.8 | 0.04 | 0.55 | −1.99 | √ | √ | √ |
| DM_5 | Fumaric acid | 116.0101 | 116.01 | 0.49 | 12.6 | 0.05 | 0.42 | −2.00 | √ | √ | √ |
| DM_6 | Propane-1,2,3-triol | 92.0473 | 92.04703 | 2.9 | 7.2 | 0.07 | 0.99 | −1.84 | √ | × | × |
| DM_7 | 2-Hydroxypropanoic acid | 90.0317 | 90.03207 | −4.07 | 12.1 | 0.04 | 0.32 | −1.00 | √ | × | × |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wahman, R.; Sauvêtre, A.; Schröder, P.; Moser, S.; Letzel, T. Untargeted Metabolomics Studies on Drug-Incubated Phragmites australis Profiles. Metabolites 2021, 11, 2. https://doi.org/10.3390/metabo11010002
Wahman R, Sauvêtre A, Schröder P, Moser S, Letzel T. Untargeted Metabolomics Studies on Drug-Incubated Phragmites australis Profiles. Metabolites. 2021; 11(1):2. https://doi.org/10.3390/metabo11010002
Chicago/Turabian StyleWahman, Rofida, Andres Sauvêtre, Peter Schröder, Stefan Moser, and Thomas Letzel. 2021. "Untargeted Metabolomics Studies on Drug-Incubated Phragmites australis Profiles" Metabolites 11, no. 1: 2. https://doi.org/10.3390/metabo11010002
APA StyleWahman, R., Sauvêtre, A., Schröder, P., Moser, S., & Letzel, T. (2021). Untargeted Metabolomics Studies on Drug-Incubated Phragmites australis Profiles. Metabolites, 11(1), 2. https://doi.org/10.3390/metabo11010002