Simultaneous Measurement of Amino Acid Enantiomers in Aged Mouse Brain Samples by LC/MS/MS Combined with Derivatization Using Nα-(5-Fluoro-2,4-dinitrophenyl)-l-leucinamide (l-FDLA)
Abstract
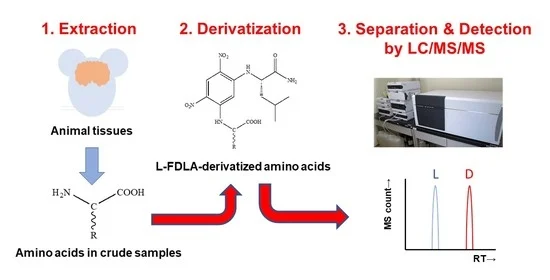
:1. Introduction
2. Results
2.1. Optimization of the Multiple Reaction Monitoring (MRM) Settings of LC/MS/MS for l-FDLA Derivatized Amino Acids
2.2. Detection of Derivatized Standard Compounds by LC/MS/MS
2.3. Detection of Amino Acid Enantiomers in Biological Samples
2.4. Validation of the Absolute Quantification of Amino Acid Enantiomers
2.5. Changes of Amino Acid Enantiomers in Aged Brains
3. Discussion
4. Materials and Methods
4.1. Reagents
4.2. Animals
4.3. Metabolite Extraction from Animal Tissues
4.4. Derivatization of Amino Acids
4.5. LC/MS/MS Condition
4.6. Quantification of Amino Acids
4.7. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Genchi, G. An overview on d-amino acids. Amino Acids 2017, 49, 1521–1533. [Google Scholar] [CrossRef] [PubMed]
- Seckler, J.M.; Lewis, S.J. Advances in d-Amino Acids in Neurological Research. Int. J. Mol. Sci. 2020, 21, 7325. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, A.; Nishikawa, T.; Hayashi, T.; Fujii, N.; Harada, K.; Oka, T.; Takahashi, K. The presence of free d-serine in rat brain. FEBS Lett. 1992, 296, 33–36. [Google Scholar] [CrossRef] [Green Version]
- Usiello, A.; Di Fiore, M.M.; De Rosa, A.; Falvo, S.; Errico, F.; Santillo, A.; Nuzzo, T.; Chieffi Baccari, G. New Evidence on the Role of d-Aspartate Metabolism in Regulating Brain and Endocrine System Physiology: From Preclinical Observations to Clinical Applications. Int. J. Mol. Sci. 2020, 21, 8718. [Google Scholar] [CrossRef]
- Ploux, E.; Freret, T.; Billard, J.M. d-serine in physiological and pathological brain aging. Biochim. Biophys. Acta Proteins Proteom. 2021, 1869, 140542. [Google Scholar] [CrossRef]
- Man, E.H.; Sandhouse, M.E.; Burg, J.; Fisher, G.H. Accumulation of d-aspartic acid with age in the human brain. Science 1983, 220, 1407–1408. [Google Scholar] [CrossRef]
- Fisher, G.H.; D’Aniello, A.; Vetere, A.; Padula, L.; Cusano, G.P.; Man, E.H. Free d-aspartate and d-alanine in normal and Alzheimer brain. Brain Res. Bull. 1991, 26, 983–985. [Google Scholar] [CrossRef]
- Dunlop, D.S.; Neidle, A.; McHale, D.; Dunlop, D.M.; Lajtha, A. The presence of free d-aspartic acid in rodents and man. Biochem. Biophys. Res. Commun. 1986, 141, 27–32. [Google Scholar] [CrossRef]
- Van Veldhoven, P.P.; Brees, C.; Mannaerts, G.P. d-Aspartate oxidase, a peroxisomal enzyme in liver of rat and man. Biochim. Biophys. Acta Gen. Subj. 1991, 1073, 203–208. [Google Scholar] [CrossRef]
- Punzo, D.; Errico, F.; Cristino, L.; Sacchi, S.; Keller, S.; Belardo, C.; Luongo, L.; Nuzzo, T.; Imperatore, R.; Florio, E.; et al. Age-Related Changes in d-Aspartate Oxidase Promoter Methylation Control Extracellular d-Aspartate Levels and Prevent Precocious Cell Death during Brain Aging. J. Neurosci. 2016, 36, 3064–3078. [Google Scholar] [CrossRef]
- Chieffi Baccari, G.; Falvo, S.; Santillo, A.; Di Giacomo Russo, F.; Di Fiore, M.M. d-Amino acids in mammalian endocrine tissues. Amino Acids 2020, 52, 1263–1273. [Google Scholar] [CrossRef] [PubMed]
- D’Aniello, A.; Di Fiore, M.M.; Fisher, G.H.; Milone, A.; Seleni, A.; D’Aniello, S.; Perna, A.F.; Ingrosso, D. Occurrence of d-aspartic acid and N-methyl-d-aspartic acid in rat neuroendocrine tissues and their role in the modulation of luteinizing hormone and growth hormone release. FASEB J. 2000, 14, 699–714. [Google Scholar] [CrossRef] [PubMed]
- Di Fiore, M.M.; Santillo, A.; Falvo, S.; Chieffi Baccari, G.; Venditti, M.; Di Giacomo Russo, F.; Lispi, M.; D’Aniello, A. Sex hormone levels in the brain of d-aspartate-treated rats. C R. Biol. 2018, 341, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Santillo, A.; Falvo, S.; Chieffi, P.; Burrone, L.; Chieffi Baccari, G.; Longobardi, S.; Di Fiore, M.M. d-aspartate affects NMDA receptor-extracellular signal-regulated kinase pathway and upregulates androgen receptor expression in the rat testis. Theriogenology 2014, 81, 744–751. [Google Scholar] [CrossRef]
- Molla, G.; Piubelli, L.; Volontè, F.; Pilone, M.S. Enzymatic detection of d-amino acids. Methods Mol. Biol. 2012, 794, 273–289. [Google Scholar] [CrossRef]
- Nagata, Y.; Akino, T.; Ohno, K. The presence of free d-amino acids in mouse tissues. Experientia 1989, 45, 330–332. [Google Scholar] [CrossRef]
- Hinkkanen, A.; Decker, K. Enzymatic determination of several d-amino acids using luminol-mediated chemiluminescence. Hoppe Seylers Z. Physiol. Chem. 1983, 364, 1549–1553. [Google Scholar] [CrossRef]
- Buck, R.H.; Krummen, K. High-performance liquid chromatographic determination of enantiomeric amino acids and amino alcohols after derivatization with o-phthaldialdehyde and various chiral mercaptans. Application to peptide hydrolysates. J. Chromatogr. 1987, 387, 255–265. [Google Scholar] [CrossRef]
- Brückner, H.; Westhauser, T.; Godel, H. Liquid chromatographic determination of d- and l-amino acids by derivatization with o-phthaldialdehyde and N-isobutyryl-L-cysteine. Applications with reference to the analysis of peptidic antibiotics, toxins, drugs and pharmaceutically used amino acids. J. Chromatogr. A 1995, 711, 201–215. [Google Scholar] [CrossRef]
- Okuma, E.; Abe, H. Simultaneous determination of d- and l-amino acids in the nervous tissues of crustaceans using precolumn derivatization with (+)-1-(9-fluorenyl)ethyl chloroformate and reversed-phase ion-pair high-performance liquid chromatography. J. Chromatogr. B Biomed. Appl. 1994, 660, 243–250. [Google Scholar] [CrossRef]
- Ishii, C.; Akita, T.; Mita, M.; Ide, T.; Hamase, K. Development of an online two-dimensional high-performance liquid chromatographic system in combination with tandem mass spectrometric detection for enantiomeric analysis of free amino acids in human physiological fluid. J. Chromatogr. A 2018, 1570, 91–98. [Google Scholar] [CrossRef] [PubMed]
- Hyun, M.H.; Jin, J.S.; Lee, W. Liquid chromatographic resolution of racemic amino acids and their derivatives on a new chiral stationary phase based on crown ether. J. Chromatogr. A 1998, 822, 155–161. [Google Scholar] [CrossRef]
- Teixeira, J.; Tiritan, M.E.; Pinto, M.M.M.; Fernandes, C. Chiral Stationary Phases for Liquid Chromatography: Recent Developments. Molecules 2019, 24, 865. [Google Scholar] [CrossRef] [Green Version]
- Lämmerhofer, M. Chiral recognition by enantioselective liquid chromatography: Mechanisms and modern chiral stationary phases. J. Chromatogr. A 2010, 1217, 814–856. [Google Scholar] [CrossRef] [PubMed]
- Ilisz, I.; Aranyi, A.; Péter, A. Chiral derivatizations applied for the separation of unusual amino acid enantiomers by liquid chromatography and related techniques. J. Chromatogr. A 2013, 1296, 119–139. [Google Scholar] [CrossRef]
- Marfey, P. Determination of d-amino acids. II. Use of a bifunctional reagent, 1,5-difluoro-2,4-dinitrobenzene. Carlsberg Res. Commun. 1984, 49, 591. [Google Scholar] [CrossRef] [Green Version]
- Ayon, N.J.; Sharma, A.D.; Gutheil, W.G. LC-MS/MS-Based Separation and Quantification of Marfey’s Reagent Derivatized Proteinogenic Amino Acid DL-Stereoisomers. J. Am. Soc. Mass Spectrom. 2019, 30, 448–458. [Google Scholar] [CrossRef]
- Fujii, K.; Ikai, Y.; Oka, H.; Suzuki, M.; Harada, K.-i. A Nonempirical Method Using LC/MS for Determination of the Absolute Configuration of Constituent Amino Acids in a Peptide: Combination of Marfey’s Method with Mass Spectrometry and Its Practical Application. Anal. Chem. 1997, 69, 5146–5151. [Google Scholar] [CrossRef]
- Papouin, T.; Ladépêche, L.; Ruel, J.; Sacchi, S.; Labasque, M.; Hanini, M.; Groc, L.; Pollegioni, L.; Mothet, J.P.; Oliet, S.H. Synaptic and extrasynaptic NMDA receptors are gated by different endogenous coagonists. Cell 2012, 150, 633–646. [Google Scholar] [CrossRef] [Green Version]
- Topo, E.; Soricelli, A.; Di Maio, A.; D’Aniello, E.; Di Fiore, M.M.; D’Aniello, A. Evidence for the involvement of d-aspartic acid in learning and memory of rat. Amino Acids 2010, 38, 1561–1569. [Google Scholar] [CrossRef]
- Suzuki, M.; Imanishi, N.; Mita, M.; Hamase, K.; Aiso, S.; Sasabe, J. Heterogeneity of d-Serine Distribution in the Human Central Nervous System. ASN Neuro 2017, 9, 1759091417713905. [Google Scholar] [CrossRef] [PubMed]
- Hamase, K.; Homma, H.; Takigawa, Y.; Fukushima, T.; Santa, T.; Imai, K. Regional distribution and postnatal changes of d-amino acids in rat brain. Biochim. Biophys. Acta 1997, 1334, 214–222. [Google Scholar] [CrossRef]
- Billard, J.M. d-Serine in the aging hippocampus. J. Pharm. Biomed. Anal. 2015, 116, 18–24. [Google Scholar] [CrossRef] [PubMed]
- Potier, B.; Turpin, F.R.; Sinet, P.M.; Rouaud, E.; Mothet, J.P.; Videau, C.; Epelbaum, J.; Dutar, P.; Billard, J.M. Contribution of the d-Serine-Dependent Pathway to the Cellular Mechanisms Underlying Cognitive Aging. Front. Aging Neurosci. 2010, 2, 1. [Google Scholar] [CrossRef] [Green Version]
- Junjaud, G.; Rouaud, E.; Turpin, F.; Mothet, J.P.; Billard, J.M. Age-related effects of the neuromodulator d-serine on neurotransmission and synaptic potentiation in the CA1 hippocampal area of the rat. J. Neurochem. 2006, 98, 1159–1166. [Google Scholar] [CrossRef]
- Einarsson, S.; Josefsson, B.; Möller, P.; Sanchez, D. Separation of amino acid enantiomers and chiral amines using precolumn derivatization with (+)-1-(9-fluorenyl)ethyl chloroformate and reversed-phase liquid chromatography. Anal. Chem. 1987, 59, 1191–1195. [Google Scholar] [CrossRef]
- Langguth, P.; Spahn, H.; Merkle, H.P. Fluorescence assay for small peptides and amino acids: High-performance liquid chromatographic determination of selected substrates using activated S-flunoxaprofen as a chiral derivatizing agent. J. Chromatogr. 1990, 528, 55–64. [Google Scholar] [CrossRef]
- Thorsén, G.; Engström, A.; Josefsson, B. Enantiomeric determination of amino compounds with high sensitivity using the chiral reagents (+)- and (-)-1-(9-anthryl)-2-propyl chloroformate. J. Chromatogr. A 1997, 786, 347–354. [Google Scholar] [CrossRef]
- Aswad, D.W. Determination of d- and L-aspartate in amino acid mixtures by high-performance liquid chromatography after derivatization with a chiral adduct of o-phthaldialdehyde. Anal. Biochem. 1984, 137, 405–409. [Google Scholar] [CrossRef]
- Brückner, H.; Wittner, R.; Godel, H. Automated enantioseparation of amino acids by derivatization with o-phthaldialdehyde and n-acylated cysteines. J. Chromatogr. 1989, 476, 73–82. [Google Scholar] [CrossRef]
- Weatherly, C.A.; Du, S.; Parpia, C.; Santos, P.T.; Hartman, A.L.; Armstrong, D.W. d-Amino Acid Levels in Perfused Mouse Brain Tissue and Blood: A Comparative Study. ACS Chem. Neurosci. 2017, 8, 1251–1261. [Google Scholar] [CrossRef] [PubMed]
- Inoue, T.; Hamase, K.; Morikawa, A.; Zaitsu, K. Determination of minute amounts of d-leucine in various brain regions of rat and mouse using column-switching high-performance liquid chromatography. J. Chromatogr. B Biomed. Sci. Appl. 2000, 744, 213–219. [Google Scholar] [CrossRef]
- Glushakov, A.V.; Dennis, D.M.; Morey, T.E.; Sumners, C.; Cucchiara, R.F.; Seubert, C.N.; Martynyuk, A.E. Specific inhibition of N-methyl-d-aspartate receptor function in rat hippocampal neurons by L-phenylalanine at concentrations observed during phenylketonuria. Mol. Psychiatry 2002, 7, 359–367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Glushakov, A.V.; Dennis, D.M.; Sumners, C.; Seubert, C.N.; Martynyuk, A.E. L-phenylalanine selectively depresses currents at glutamatergic excitatory synapses. J. Neurosci. Res. 2003, 72, 116–124. [Google Scholar] [CrossRef]
- Irukayama-Tomobe, Y.; Tanaka, H.; Yokomizo, T.; Hashidate-Yoshida, T.; Yanagisawa, M.; Sakurai, T. Aromatic d-amino acids act as chemoattractant factors for human leukocytes through a G protein-coupled receptor, GPR109B. Proc. Natl. Acad. Sci. USA 2009, 106, 3930–3934. [Google Scholar] [CrossRef] [Green Version]
- Ariyoshi, M.; Katane, M.; Hamase, K.; Miyoshi, Y.; Nakane, M.; Hoshino, A.; Okawa, Y.; Mita, Y.; Kaimoto, S.; Uchihashi, M.; et al. (D)-Glutamate is metabolized in the heart mitochondria. Sci. Rep. 2017, 7, 43911. [Google Scholar] [CrossRef] [Green Version]
- Topo, E.; Fisher, G.; Sorricelli, A.; Errico, F.; Usiello, A.; D’Aniello, A. Thyroid hormones and d-aspartic acid, d-aspartate oxidase, d-aspartate racemase, H2O2, and ROS in rats and mice. Chem. Biodivers. 2010, 7, 1467–1478. [Google Scholar] [CrossRef]
- Hashimoto, A.; Nishikawa, T.; Oka, T.; Hayashi, T.; Takahashi, K. Widespread distribution of free d-aspartate in rat periphery. FEBS Lett. 1993, 331, 4–8. [Google Scholar] [CrossRef] [Green Version]
- Lee, C.J.; Qiu, T.A.; Sweedler, J.V. d-Alanine: Distribution, origin, physiological relevance, and implications in disease. Biochim. Biophys. Acta Proteins Proteom. 2020, 1868, 140482. [Google Scholar] [CrossRef]
- Kimura, T.; Hamase, K.; Miyoshi, Y.; Yamamoto, R.; Yasuda, K.; Mita, M.; Rakugi, H.; Hayashi, T.; Isaka, Y. Chiral amino acid metabolomics for novel biomarker screening in the prognosis of chronic kidney disease. Sci. Rep. 2016, 6, 26137. [Google Scholar] [CrossRef]
- Yaku, K.; Okabe, K.; Gulshan, M.; Takatsu, K.; Okamoto, H.; Nakagawa, T. Metabolism and biochemical properties of nicotinamide adenine dinucleotide (NAD) analogs, nicotinamide guanine dinucleotide (NGD) and nicotinamide hypoxanthine dinucleotide (NHD). Sci. Rep. 2019, 9, 13102. [Google Scholar] [CrossRef] [PubMed]
- Yaku, K.; Okabe, K.; Nakagawa, T. Simultaneous measurement of NAD metabolome in aged mice tissue using liquid chromatography tandem-mass spectrometry. Biomed. Chromatogr. 2018, 32, e4205. [Google Scholar] [CrossRef] [PubMed]





| Monoisotopic Mass | Retention Time | ||||||
|---|---|---|---|---|---|---|---|
| Before Derivatization | After Derivatization | Precursor Ion (m/z) | Product Ion (m/z) | Collision Energy (eV) | l-AA (min) | d-AA (min) | |
| Ala | 89.0477 | 383.1441 | 384 | 339 | 6 | 17.0 | 18.9 |
| Asn | 132.0535 | 426.1499 | 427 | 382 | 8 | 16.0 | 17.0 |
| Asp | 133.0375 | 427.1339 | 428 | 383 | 6 | 14.0 | 14.9 |
| Gln | 146.0691 | 440.1656 | 441 | 334 | 10 | 16.7 | 17.3 |
| Glu | 147.0532 | 441.1496 | 442 | 397 | 8 | 15.2 | 16.0 |
| Leu | 131.0946 | 425.1910 | 426 | 380 | 4 | 17.9 | 22.4 |
| Met | 149.0510 | 443.1475 | 444 | 354 | 8 | 17.4 | 20.2 |
| Phe | 165.0790 | 459.1754 | 460 | 415 | 8 | 17.7 | 21.0 |
| Pro | 115.0633 | 409.1597 | 410 | 365 | 10 | 16.8 | 18.5 |
| Ser | 105.0426 | 399.1390 | 400 | 355 | 6 | 16.4 | 17.1 |
| Young | Old | |||
|---|---|---|---|---|
| l-Asn (µmol/g tissue) | Cerebellum | 0.107 ± 0.018 | 0.105 ± 0.024 | n.s. |
| Cortex | 0.135 ± 0.038 | 0.131 ± 0.016 | n.s. | |
| Hippocampus | 0.150 ± 0.015 | 0.113 ± 0.019 | p < 0.05 | |
| Thalamus | 0.130 ± 0.025 | 0.122 ± 0.022 | n.s. | |
| d-Asn (µmol/g tissue) | Cerebellum | 0.000725 ± 0.000111 | 0.00100 ± 0.00047 | n.s. |
| Cortex | 0.000859 ± 0.000246 | 0.00103 ± 0.00024 | n.s. | |
| Hippocampus | 0.00107 ± 0.00006 | 0.000893 ± 0.000230 | n.s. | |
| Thalamus | 0.000625 ± 0.000136 | 0.000620 ± 0.000222 | n.s. | |
| l-Asp (µmol/g tissue) | Cerebellum | 11.3 ± 2.3 | 9.39 ± 2.28 | n.s. |
| Cortex | 7.02 ± 2.04 | 7.79 ± 0.44 | n.s. | |
| Hippocampus | 9.54 ± 1.04 | 7.37 ± 1.24 | p < 0.05 | |
| Thalamus | 7.88 ± 0.74 | 6.73 ± 1.51 | n.s. | |
| d-Asp (µmol/g tissue) | Cerebellum | 0.0126 ± 0.0053 | 0.0147 ± 0.0043 | n.s. |
| Cortex | 0.0675 ± 0.0253 | 0.0608 ± 0.0062 | n.s. | |
| Hippocampus | 0.0825 ± 0.0130 | 0.0560 ± 0.0121 | p < 0.05 | |
| Thalamus | 0.0422 ± 0.0195 | 0.0311 ± 0.0060 | n.s. | |
| l-Phe (µmol/g tissue) | Cerebellum | 0.107 ± 0.009 | 0.119 ± 0.034 | n.s. |
| Cortex | 0.0996 ± 0.0231 | 0.112 ± 0.019 | n.s. | |
| Hippocampus | 0.129 ± 0.012 | 0.127 ± 0.026 | n.s. | |
| Thalamus | 0.0966 ± 0.0212 | 0.101 ± 0.032 | n.s. | |
| d-Phe (µmol/g tissue) | Cerebellum | 0.287 ± 0.096 | 0.221 ± 0.041 | n.s. |
| Cortex | 0.246 ± 0.059 | 0.201 ± 0.057 | n.s. | |
| Hippocampus | 0.207 ± 0.069 | 0.209 ± 0.057 | n.s. | |
| Thalamus | 0.211 ± 0.015 | 0.241 ± 0.015 | p < 0.05 | |
| l-Ser (µmol/g tissue) | Cerebellum | 0.983 ± 0.114 | 1.06 ± 0.29 | n.s. |
| Cortex | 1.31 ± 0.34 | 1.30 ± 0.07 | n.s. | |
| Hippocampus | 1.56 ± 0.09 | 1.32 ± 0.18 | p < 0.05 | |
| Thalamus | 0.896 ± 0.235 | 0.819 ± 0.139 | n.s. | |
| d-Ser (µmol/g tissue) | Cerebellum | 0.0285 ± 0.0158 | 0.0211 ± 0.0080 | n.s. |
| Cortex | 0.609 ± 0.156 | 0.580 ± 0.048 | n.s. | |
| Hippocampus | 0.646 ± 0.041 | 0.526 ± 0.060 | p < 0.01 | |
| Thalamus | 0.313 ± 0.109 | 0.279 ± 0.036 | n.s. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yamamoto, T.; Yaku, K.; Nakagawa, T. Simultaneous Measurement of Amino Acid Enantiomers in Aged Mouse Brain Samples by LC/MS/MS Combined with Derivatization Using Nα-(5-Fluoro-2,4-dinitrophenyl)-l-leucinamide (l-FDLA). Metabolites 2021, 11, 57. https://doi.org/10.3390/metabo11010057
Yamamoto T, Yaku K, Nakagawa T. Simultaneous Measurement of Amino Acid Enantiomers in Aged Mouse Brain Samples by LC/MS/MS Combined with Derivatization Using Nα-(5-Fluoro-2,4-dinitrophenyl)-l-leucinamide (l-FDLA). Metabolites. 2021; 11(1):57. https://doi.org/10.3390/metabo11010057
Chicago/Turabian StyleYamamoto, Taiji, Keisuke Yaku, and Takashi Nakagawa. 2021. "Simultaneous Measurement of Amino Acid Enantiomers in Aged Mouse Brain Samples by LC/MS/MS Combined with Derivatization Using Nα-(5-Fluoro-2,4-dinitrophenyl)-l-leucinamide (l-FDLA)" Metabolites 11, no. 1: 57. https://doi.org/10.3390/metabo11010057
APA StyleYamamoto, T., Yaku, K., & Nakagawa, T. (2021). Simultaneous Measurement of Amino Acid Enantiomers in Aged Mouse Brain Samples by LC/MS/MS Combined with Derivatization Using Nα-(5-Fluoro-2,4-dinitrophenyl)-l-leucinamide (l-FDLA). Metabolites, 11(1), 57. https://doi.org/10.3390/metabo11010057