Methodology for Single Bee and Bee Brain 1H-NMR Metabolomics
Abstract
:1. Introduction
2. Results and Discussion
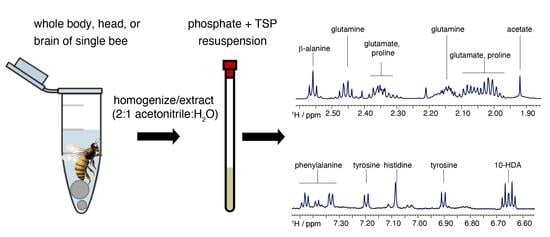
2.1. Extraction Methodology
2.2. Honeybee Brain
2.3. Low Field NMR
2.4. Other Bees and Pollinators: Carpenter Bee
3. Materials and Methods
3.1. Homogenization
3.2. Chloroform/Methanol Extraction
3.3. Acetonitrile Extraction
3.4. NMR Spectroscopy
3.5. Brain Dissection
3.6. Hive Management
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kang, Y.; Blanco, K.; Davis, T.; Wang, Y.; DeGrandi-Hoffman, G. Disease dynamics of honeybees with Varroa destructor as parasite and virus vector. Math. Biosci. 2016, 275, 71–92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Williams, G.R.; Tarpy, D.R.; vanEngelsdorp, D.; Chauzat, M.; Cox-Foster, D.; Delaplane, K.S.; Neumann, P.; Pettis, J.S.; Rogers, R.E.L.; Shutler, D. Colony Collapse Disorder in context. Bioessays 2010, 32, 845–846. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- vanEngelsdorp, D.; Evans, J.D.; Saegerman, C.; Mullin, C.; Haubruge, E.; Nguyen, B.K.; Frazier, M.; Frazier, J.; Cox-Foster, D.; Chen, Y.; et al. Colony Collapse Disorder: A Descriptive Study. PLoS ONE 2009, 4, e6481. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekaran, S.; Rittschof, C.C.; Djukovic, D.; Gu, H.; Raftery, D.; Price, N.D.; Robinson, G.E. Aggression is associated with aerobic glycolysis in the honey bee brain1. Genes Brain Behav. 2015, 14, 158–166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pratavieira, M.; da Silva Menegasso, A.R.; Roat, T.; Malaspina, O.; Palma, M.S. In Situ Metabolomics of the Honeybee Brain: The Metabolism of l-Arginine through the Polyamine Pathway in the Proboscis Extension Response (PER). J. Proteome Res. 2020, 19, 832–844. [Google Scholar] [CrossRef] [PubMed]
- Giacometti, R.; Jacobi, V.; Kronberg, F.; Panagos, C.; Edison, A.S.; Zavala, J.A. Digestive activity and organic compounds of Nezara viridula watery saliva induce defensive soybean seed responses. Sci. Rep. 2020, 10, 15468. [Google Scholar] [CrossRef]
- Riach, A.C.; Perera, M.V.L.; Florance, H.V.; Robinson, L.A.; Penfield, S.D.; Hill, J.K. Metabolic fingerprints reveal how an insect metabolome is affected by different larval host plant species. Arthropod Plant Interact. 2019, 13, 571–579. [Google Scholar] [CrossRef]
- Snart, C.J.P.; Hardy, I.C.W.; Barrett, D.A. Entometabolomics: Applications of modern analytical techniques to insect studies. Entomol. Exp. Appl. 2015, 155, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Shi, T.; Burton, S.; Wang, Y.; Xu, S.; Zhang, W.; Yu, L. Metabolomic analysis of honey bee, Apis mellifera L. response to thiacloprid. Pestic Biochem. Physiol. 2018, 152, 17–23. [Google Scholar] [CrossRef]
- Wu, J.; Zhou, C.; Wu, P.; Xu, J.; Guo, Y.; Xue, F.; Getachew, A.; Xu, S. Brain metabolomic profiling of eastern honey bee (Apis cerana) infested with the mite Varroa destructor. PLoS ONE 2017, 12, e0175573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rothman Jason, A.; Laura, L.; Kirkwood Jay, S.; McFrederick Quinn, S.; Stabb Eric, V. Cadmium and Selenate Exposure Affects the Honey Bee Microbiome and Metabolome, and Bee-Associated Bacteria Show Potential for Bioaccumulation. Appl. Environ. Microbiol. 2019, 85, 1411. [Google Scholar] [CrossRef] [Green Version]
- Dossey, A.T.; Walse, S.S.; Rocca, J.R.; Edison, A.S. Single-Insect NMR: A New Tool to Probe Chemical Biodiversity. ACS Chem. Biol. 2006, 1, 511–514. [Google Scholar] [CrossRef] [PubMed]
- Deng, M.; Lin, X.; Lin, Q.; Wen, D.; Zhang, M.; Wang, X.; Gao, H.; Xu, J. A 1H-NMR Based Study on Hemolymph Metabolomics in Eri Silkworm after Oral Administration of 1-Deoxynojirimycin. PLoS ONE 2015, 10, e0131696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ardalani, H.; Vidkjær, N.H.; Kryger, P.; Fiehn, O.; Fomsgaard, I.S. Metabolomics unveils the influence of dietary phytochemicals on residual pesticide concentrations in honey bees. Env. Int. 2021, 152, 106503. [Google Scholar] [CrossRef] [PubMed]
- Pizzorno, M.C.; Field, K.; Kobokovich, A.L.; Martin, P.L.; Gupta, R.A.; Mammone, R.; Rovnyak, D.; Capaldi, E.A. Transcriptomic Responses of the Honey Bee Brain to Infection with Deformed Wing Virus. Viruses 2021, 13, 287. [Google Scholar] [CrossRef] [PubMed]
- Tackenberg, M.C.; Giannoni-Guzmán, M.A.; Sanchez-Perez, E.; Doll, C.A.; Agosto-Rivera, J.L.; Broadie, K.; Moore, D.; McMahon, D.G. Neonicotinoids disrupt circadian rhythms and sleep in honey bees. Sci. Rep. 2020, 10, 17929. [Google Scholar] [CrossRef] [PubMed]
- Cabirol, A.; Haase, A. The Neurophysiological Bases of the Impact of Neonicotinoid Pesticides on the Behaviour of Honeybees. Insects 2019, 10, 344. [Google Scholar] [CrossRef] [Green Version]
- Ardalani, H.; Vidkjær, N.H.; Laursen, B.B.; Kryger, P.; Fomsgaard, I.S. Dietary quercetin impacts the concentration of pesticides in honey bees. Chemosphere 2021, 262, 127848. [Google Scholar] [CrossRef] [PubMed]
- Wilson, M.B.; Spivak, M.; Hegeman, A.D.; Rendahl, A.; Cohen, J.D. Metabolomics Reveals the Origins of Antimicrobial Plant Resins Collected by Honey Bees. PLoS ONE 2013, 8, e77512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kešnerová, L.; Emery, O.; Troilo, M.; Liberti, J.; Erkosar, B.; Engel, P. Gut microbiota structure differs between honeybees in winter and summer. ISME J. 2020, 14, 801–814. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kešnerová, L.; Mars, R.A.T.; Ellegaard, K.M.; Troilo, M.; Sauer, U.; Engel, P. Disentangling metabolic functions of bacteria in the honey bee gut. PLoS Biol. 2017, 15, e2003467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yong, C.; Muhammad, S.A.; Aziz, F.A.; Nasir, F.I.; Mustafa, M.Z.; Ibrahim, B.; Kelly, S.D.; Cannavan, A.; Seow, E. Detecting adulteration of stingless bee honey using untargeted 1H NMR metabolomics with chemometrics. Food Chem. 2022, 368, 130808. [Google Scholar] [CrossRef] [PubMed]
- Mazzei, P.; Piccolo, A.; Brescia, M.; Caprio, E. Assessment of geographical origin and production period of royal jelly by NMR metabolomics. Chem. Biol. Technol. Agric. 2020, 7, 24. [Google Scholar] [CrossRef]
- Wang, L.; Meeus, I.; Rombouts, C.; Van Meulebroek, L.; Vanhaecke, L.; Smagghe, G. Metabolomics-based biomarker discovery for bee health monitoring: A proof of concept study concerning nutritional stress in Bombus terrestris. Sci. Rep. 2019, 9, 11423. [Google Scholar] [CrossRef]
- Grainger, M.N.C.; Hewitt, N.; French, A.D. Optimised approach for small mass sample preparation and elemental analysis of bees and bee products by inductively coupled plasma mass spectrometry. Talanta 2020, 214, 120858. [Google Scholar] [CrossRef] [PubMed]
- Bligh, E.G.; Dyer, W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef]
- Nagana Gowda, G.A.; Gowda, Y.N.; Raftery, D. Expanding the limits of human blood metabolite quantitation using NMR spectroscopy. Anal. Chem. 2015, 87, 706–715. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagana Gowda, G.A.; Raftery, D. Quantitating metabolites in protein precipitated serum using NMR spectroscopy. Anal. Chem. 2014, 86, 5433–5440. [Google Scholar] [CrossRef]
- Miele, M.M.; Irving, B.A.; Wenrich, B.R.; Martin, P.L. Reproducibility and Stability of Aqueous Metabolite Levels in Extracted Serum by NMR Spectroscopy. Curr. Metab. 2017, 5, 45–54. [Google Scholar] [CrossRef]
- Nagana Gowda, G.A.; Hong, N.N.; Raftery, D. Evaluation of Fumaric Acid and Maleic Acid as Internal Standards for NMR Analysis of Protein Precipitated Plasma, Serum, and Whole Blood. Anal. Chem. 2021, 93, 3233–3240. [Google Scholar] [CrossRef]
- Yang, X.; Yang, S.; Wang, R. Comparative proteomic analysis provides insight into 10-hydroxy-2-decenoic acid biosynthesis in honey bee workers. Amino Acids 2017, 49, 1177–1192. [Google Scholar] [CrossRef]
- Engels, W.; Rosenkranz, P.; Adler, A.; Taghizadeh, T.; Lübke, G.; Francke, W. Mandibular Gland Volatiles and Their Ontogenetic Patterns in Queen Honey Bees, Apis mellifera carnica. J. Insect Physiol. 1997, 43, 307–313. [Google Scholar] [CrossRef]
- Milone, J.P.; Chakrabarti, P.; Sagili, R.R.; Tarpy, D.R. Colony-level pesticide exposure affects honey bee (Apis mellifera L.) royal jelly production and nutritional composition. Chemosphere 2021, 263, 128183. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Robinson, G.E.; Tobe, S.S.; Yagi, K.J.; Strambi, C.; Strambi, A.; Stay, B. Hormonal regulation of behavioural development in the honey bee is based on changes in the rate of juvenile hormone biosynthesis. J. Insect Physiol. 1991, 37, 733–741. [Google Scholar] [CrossRef]
- Mustard, J.A.; Jones, L.; Wright, G.A. GABA signaling affects motor function in the honey bee. J. Insect Physiol. 2020, 120, 103989. [Google Scholar] [CrossRef]
- Pan, Z.; Raftery, D. Comparing and combining NMR spectroscopy and mass spectrometry in metabolomics. Anal. Bioanal. Chem. 2007, 387, 525–527. [Google Scholar] [CrossRef]
- van Beek, T.A. Low-field benchtop NMR spectroscopy: Status and prospects in natural product analysis. Phytochem. Anal. 2021, 32, 24–37. [Google Scholar] [CrossRef] [Green Version]
- Michal, C.A. Low-cost low-field NMR and MRI: Instrumentation and applications. J. Magn. Reson. 2020, 319, 106800. [Google Scholar] [CrossRef]








Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
McDevitt, J.C.; Gupta, R.A.; Dickinson, S.G.; Martin, P.L.; Rieuthavorn, J.; Freund, A.; Pizzorno, M.C.; Capaldi, E.A.; Rovnyak, D. Methodology for Single Bee and Bee Brain 1H-NMR Metabolomics. Metabolites 2021, 11, 864. https://doi.org/10.3390/metabo11120864
McDevitt JC, Gupta RA, Dickinson SG, Martin PL, Rieuthavorn J, Freund A, Pizzorno MC, Capaldi EA, Rovnyak D. Methodology for Single Bee and Bee Brain 1H-NMR Metabolomics. Metabolites. 2021; 11(12):864. https://doi.org/10.3390/metabo11120864
Chicago/Turabian StyleMcDevitt, Jayne C., Riju A. Gupta, Sydney G. Dickinson, Phillip L. Martin, Jean Rieuthavorn, Amy Freund, Marie C. Pizzorno, Elizabeth A. Capaldi, and David Rovnyak. 2021. "Methodology for Single Bee and Bee Brain 1H-NMR Metabolomics" Metabolites 11, no. 12: 864. https://doi.org/10.3390/metabo11120864
APA StyleMcDevitt, J. C., Gupta, R. A., Dickinson, S. G., Martin, P. L., Rieuthavorn, J., Freund, A., Pizzorno, M. C., Capaldi, E. A., & Rovnyak, D. (2021). Methodology for Single Bee and Bee Brain 1H-NMR Metabolomics. Metabolites, 11(12), 864. https://doi.org/10.3390/metabo11120864