Plant Metabolites Involved in the Differential Development of a Heliantheae-Specialist Insect
Abstract
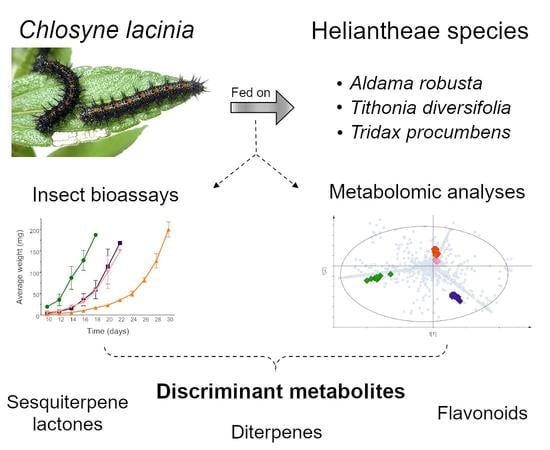
:1. Introduction
2. Results
2.1. Metabolomic Analysis
2.2. C. lacinia Development
3. Discussion
4. Materials and Methods
4.1. Experimental Design
4.2. Insects and Diets
4.3. C. lacinia Development
4.4. Metabolomic Analysis
4.4.1. Sample Preparation
4.4.2. LC-MS Analyses
4.4.3. GC-MS Analyses
4.4.4. Data Processing
4.4.5. Multivariate Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Additional Information
References
- Wilson, J.K.; Ruiz, L.; Duarte, J.; Davidowitz, G. The nutritional landscape of host plants for a specialist insect herbivore. Ecol. Evol. 2019, 9, 13104–13113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Forister, M.L.; Novotny, V.; Panorska, A.K.; Baje, L.; Basset, Y.; Butterill, P.T.; Cizek, L.; Coley, P.D.; Dem, F.; Diniz, I.R.; et al. The global distribution of diet breadth in insect herbivores. Proc. Natl. Acad. Sci. USA 2015, 112, 442–447. [Google Scholar] [CrossRef] [Green Version]
- Burkepile, D.E.; Parker, J.D. Recent advances in plant-herbivore interactions. F1000 Res. 2017, 6, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bruce, T.J.A. Interplay between insects and plants: Dynamic and complex interactions that have coevolved over millions of years but act in milliseconds. J. Exp. Bot. 2015, 66, 455–465. [Google Scholar] [CrossRef] [PubMed]
- Maag, D.; Erb, M.; Glauser, G. Metabolomics in plant-herbivore interactions: Challenges and applications. Entomol. Exp. Appl. 2015, 157, 18–29. [Google Scholar] [CrossRef] [Green Version]
- Nagler, M.; Nägele, T.; Gilli, C.; Fragner, L.; Korte, A.; Platzer, A.; Farlow, A.; Nordborg, M.; Weckwerth, W. Eco-metabolomics and metabolic modeling: Making the leap from model systems in the lab to native populations in the field. Front. Plant Sci. 2018, 9, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Peters, K.; Worrich, A.; Weinhold, A.; Alka, O.; Balcke, G.; Birkemeyer, C.; Bruelheide, H.; Calf, O.W.; Dietz, S.; Dührkop, K.; et al. Current challenges in plant eco-metabolomics. Int. J. Mol. Sci. 2018, 19, 1385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sardans, J.; Gargallo-Garriga, A.; Urban, O.; Klem, K.; Walker, T.W.N.; Holub, P.; Janssens, I.A.; Peñuelas, J. Ecometabolomics for a better understanding of plant responses and acclimation to abiotic factors linked to global change. Metabolites 2020, 10, 239. [Google Scholar] [CrossRef]
- Rivas-Ubach, A.; Peñuelas, J.; Hódar, J.A.; Oravec, M.; Paša-Tolić, L.; Urban, O.; Sardans, J. We are what we eat: A stoichiometric and ecometabolomic study of caterpillars feeding on two pine subspecies of Pinus sylvestris. Int. J. Mol. Sci. 2019, 20, 59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schoonhoven, L.M.; van Loon, J.J.A.; Dicke, M. Insect-Plant Biology, 2nd ed.; Oxford University Press: New York, NY, USA, 2005. [Google Scholar]
- Behmer, S.T. Insect herbivore nutrient regulation. Annu. Rev. Entomol. 2009, 54, 165–187. [Google Scholar] [CrossRef]
- Raubenheimer, D.; Simpson, S.J. Integrative models of nutrient balancing: Application to insects and vertebrates. Nutr. Res. Rev. 1997, 10, 151–179. [Google Scholar] [CrossRef] [Green Version]
- Simpson, S.J.; Sibly, R.M.; Lee, K.P.; Behmer, S.T.; Raubenheimer, D. Optimal foraging when regulating intake of multiple nutrients. Anim. Behav. 2004, 68, 1299–1311. [Google Scholar] [CrossRef]
- Metspalu, L.; Kruus, E.; Jõgar, K.; Kuusik, A.; Williams, I.H.; Veromann, E.; Luik, A.; Ploomi, A.; Hiiesaar, K.; Kivimägi, I.; et al. Larval food plants can regulate the cabbage moth, Mamestra brassicae population. Bull. Insectology 2013, 66, 93–101. [Google Scholar]
- Colasurdo, N.; Gélinas, Y.; Despland, E. Larval nutrition affects life history traits in a capital breeding mothi. J. Exp. Biol. 2009, 212, 1794–1800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neck, R.W. Foodplant ecology of the butterfly Chlosyne lacinia (Geyer) (Nymphalidae). I. Larval foodplants. J. Lepid. Soc. 1973, 27, 22–33. [Google Scholar]
- Drummond, B.A., III; Bush, G.L.; Emmel, T.C. The biology and laboratory culture of Chlosyne lacinia Geyer (Nymphalidae). J. Lepid. Soc. 1970, 24, 135–142. [Google Scholar]
- Paula, D.P.; Teixeira, M.M.; Timbó, R.V.; Ferreira, L.A.; Andrade, I.; Suji, E.R.; Pires, C.S.S.; Fontes, E.M.G. Metodologia de Criação em Laboratório do Ciclo Completo da Lagarta-do-Girassol Chlosyne Lacinia (Lepidoptera:Nymphalidae); Embrapa Recursos Genéticos e Biotecnologia: Brasilia, Brasil, 2009. [Google Scholar]
- Allen, T.J.; Brock, J.P.; Glassberg, J. Caterpillars in the Field and Garden: A Field Guide to the Butterfly Caterpillars of North America; Oxford University Press: New York, NY, USA, 2005; ISBN 9780195183719. [Google Scholar]
- Glassberg, J. A Swift Guide to Butterflies of Mexico and Central America; Princeton University Press: Princeton, NJ, USA, 2017; ISBN 9788578110796. [Google Scholar]
- Lopes-da-Silva, M.; Casagrande, M.M. Color polymorphism and allele frequency in a Brazilian population of the sunflower caterpillar Chlosyne lacinia saundersi (Doubleday) (Lepidoptera:Nymphalidae). Neotrop. Entomol. 2003, 32, 159–161. [Google Scholar] [CrossRef]
- Ambrósio, S.R.; Oki, Y.; Heleno, V.C.G.; Chaves, J.S.; Nascimento, P.G.B.D.; Lichston, J.E.; Constantino, M.G.; Varanda, E.M.; Da Costa, F.B. Constituents of glandular trichomes of Tithonia diversifolia: Relationships to herbivory and antifeedant activity. Phytochemistry 2008, 69, 2052–2060. [Google Scholar] [CrossRef]
- Martucci, M.E.P.; Gobbo-Neto, L. Differential secondary metabolite accumulation and performance of Chlosyne lacinia fed with Tithonia diversifolia or Vernonia polyanthes. Biochem. Syst. Ecol. 2016, 68, 156–162. [Google Scholar] [CrossRef]
- Funk, V.A.; Susanna, A.; Stuessy, T.F.; Bayer, R.J. Systematics, Evolution, and Biogeography of Compositae; International Association for Plant Taxonomy: Viena, Austria, 2009; ISBN 9783950175431. [Google Scholar]
- Chagas-Paula, D.A.; Oliveira, R.B.; Rocha, B.A.; Da Costa, F.B. Ethnobotany, chemistry, and biological activities of the genus Tithonia (Asteraceae). Chem. Biodivers. 2012, 9, 210–235. [Google Scholar] [CrossRef] [PubMed]
- Ajao, A.A.; Moteetee, A.N. Tithonia diversifolia (Hemsl) A. Gray. (Asteraceae:Heliantheae), an invasive plant of significant ethnopharmacological importance: A review. S. Afr. J. Bot. 2017, 113, 396–403. [Google Scholar] [CrossRef]
- Tagne, A.M.; Marino, F.; Cosentino, M. Tithonia diversifolia (Hemsl.) A. Gray as a medicinal plant: A comprehensive review of its ethnopharmacology, phytochemistry, pharmacotoxicology and clinical relevance. J. Ethnopharmacol. 2018, 220, 94–116. [Google Scholar] [CrossRef]
- Da Costa, F.B.; Vichnewski, W.; Herz, W. Constituents of Viguiera aspillioides and V. robusta. Biochem. Syst. Ecol. 1996, 24, 585–587. [Google Scholar] [CrossRef]
- Da Costa, F.B.; Scharr, K.; Arakawa, N.S.; Schilling, E.E.; Spring, O. Infraspecific variation in the chemistry of glandular trichomes of two Brazilian Viguiera species (Heliantheae; Asteraceae). J. Braz. Chem. Soc. 2001, 12, 403–407. [Google Scholar] [CrossRef] [Green Version]
- Ikewuchi, C.C.; Ikewuchi, J.C.; Ifeanacho, M.O. Phytochemical composition of Tridax procumbens Linn leaves: Potential as a functional food. Food Nutr. Sci. 2015, 6, 992–1004. [Google Scholar]
- Chen, W.H.; Ma, X.M.; Wu, Q.X.; Shi, Y.P. Chemical-constituent diversity of Tridax procumbens. Can. J. Chem. 2008, 86, 892–898. [Google Scholar] [CrossRef]
- Mecina, G.F.; Chia, M.A.; Cordeiro-Araújo, M.K.; Bittencourt-Oliveira, M.d.C.; Rosa, M.V.; Torres, A.; Molinillo, J.M.G.; Macías, F.A.; da Silva, R.M.G. Effect of flavonoids isolated from Tridax procumbens on the growth and toxin production of Microcystis aeruginos. Aquat. Toxicol. 2019, 211, 81–91. [Google Scholar] [CrossRef]
- Wink, M. Plant secondary metabolites modulate insect behavior-steps toward addiction? Front. Physiol. 2018, 9, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mello, M.O.; Silva-Filho, M.C. Plant-insect interactions: An evolutionary arms race between two distinct defense mechanisms. Braz. J. Plant Physiol. 2002, 14, 71–81. [Google Scholar] [CrossRef]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beger, R.; Beale, M.H.; Daykin, C.A.; Fan, T.W.M.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed minimum reporting standards for chemical analysis: Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blaženović, I.; Kind, T.; Ji, J.; Fiehn, O. Software tools and approaches for compound identification of LC-MS/MS data in metabolomics. Metabolites 2018, 8, 31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Isman, M.B. Toxicity and tolerance of sesquiterpene lactones in the migratory grasshopper, Melanoplus sanguinipes (Acrididae). Pestic. Biochem. Physiol. 1985, 24, 348–354. [Google Scholar] [CrossRef]
- Gallon, M.E.; Silva-Junior, E.A.; Amaral, J.G.; Lopes, N.P.; Gobbo-Neto, L. Natural products diversity in plant-insect interaction between Tithonia diversifolia (Asteraceae) and Chlosyne lacinia (Nymphalidae). Molecules 2019, 24, 3118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fuchs, G.; Boll, M.; Heider, J. Microbial degradation of aromatic compounds- from one strategy to four. Nat. Rev. Microbiol. 2011, 9, 803–816. [Google Scholar] [CrossRef]
- Nešvera, J.; Rucká, L.; Pátek, M. Catabolism of phenol and its derivatives in bacteria: Genes, their regulation, and use in the biodegradation of toxic pollutants. Adv. Appl. Microbiol. 2015, 93, 107–160. [Google Scholar]
- Dudareva, N.; Negre, F.; Nagegowda, D.A.; Orlova, I. Plant volatiles: Recent advances and future perspectives. Crit. Rev. Plant Sci. 2006, 25, 417–440. [Google Scholar] [CrossRef]
- Mori, K. Chemical Synthesis of Hormones, Pheromones and other Bioregulators; John Wiley & Sons: Hoboken, NJ, USA, 2010; ISBN 9780470697245. [Google Scholar]
- Havaux, M. Carotenoid oxidation products as stress signals in plants. Plant J. 2013, 79, 597–606. [Google Scholar] [CrossRef]
- Owuor, P.O. TEA Chemistry. Encycl. Food Sci. Nutr. 2003, 5743–5752. [Google Scholar]
- Berasategui, A.; Salem, H.; Paetz, C.; Santoro, M.; Gershenzon, J.; Kaltenpoth, M.; Schmidt, A. Gut microbiota of the pine weevil degrades conifer diterpenes and increases insect fitness. Mol. Ecol. 2017, 26, 4099–4110. [Google Scholar] [CrossRef] [PubMed]
- De Sousa, I.P.; Teixeira, M.V.S.; Furtado, N.A.J.C. An overview of biotransformation and toxicity of diterpenes. Molecules 2018, 23, 1387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raubenheimer, D.; Simpson, S.J. Nutritional ecology and foraging theory. Curr. Opin. Insect Sci. 2018, 27, 38–45. [Google Scholar] [CrossRef]
- Simmonds, M.S.J. Importance of flavonoids in insect-plant interactions: Feeding and oviposition. Phytochemistry 2001, 56, 245–252. [Google Scholar] [CrossRef]
- Mierziak, J.; Kostyn, K.; Kulma, A. Flavonoids as important molecules of plant interactions with the environment. Molecules 2014, 19, 16240–16265. [Google Scholar] [CrossRef]
- Murota, K.; Nakamura, Y.; Uehara, M. Flavonoid metabolism: The interaction of metabolites and gut microbiota. Biosci. Biotechnol. Biochem. 2018, 82, 600–610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Svoboda, J.A.; Feldlaufer, M.F. Neutral sterol metabolism in insects. Lipids 1991, 26, 614–618. [Google Scholar] [CrossRef]
- Janson, E.M.; Grebenok, R.J.; Behmer, S.T.; Abbot, P. Same host-plant, different sterols: Variation in sterol metabolism in an insect herbivore community. J. Chem. Ecol. 2009, 35, 1309–1319. [Google Scholar] [CrossRef]
- González-Coloma, A.; López-Balboa, C.; Santana, O.; Reina, M.; Fraga, B.M. Triterpene-based plant defenses. Phytochem. Rev. 2011, 10, 245–260. [Google Scholar] [CrossRef] [Green Version]
- Arrese, E.L.; Soulages, J.L. Insect fat body: Energy, metabolism, and regulation. Annu. Rev. Entomol. 2010, 55, 207–225. [Google Scholar] [CrossRef] [Green Version]
- Stanley, D.W.; Keddie, B.A.; Volkman, L.E. Fatty acid composition of whole bodies, specific tissues and cell lines of two lepidopteran insects. Comp. Biochem. Physiol. 1986, 85, 369–373. [Google Scholar]
- Sinclair, B.J.; Marshall, K.E. The many roles of fats in overwintering insects. J. Exp. Biol. 2018, 221. [Google Scholar] [CrossRef] [Green Version]
- Bober, R.; Rafaeli, A. Gene-silencing reveals the functional significance of pheromone biosynthesis activating neuropeptide receptor (PBAN-R) in a male moth. Proc. Natl. Acad. Sci. USA 2010, 107, 16858–16862. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nation, J.L. Semiochemicals. In Insect Physiology and Biochemistry; CRC Press: Boca Raton, FL, USA, 2016; pp. 523–560. [Google Scholar]
- Kasten, P.J.; Precetti, A.A.C.M.; Parra, J.R.P. Dados biológicos comparativos de Spodoptera frugiperda em duas dietas artificiais e substrato natural. Rev. Agric. 1978, 53, 68–78. [Google Scholar]
- Cohen, A.C. Insect Diets: Science and Technology; CRC Press: Boca Raton, FL, USA, 2004; ISBN 9780849315770. [Google Scholar]





| ID | Rt | Mode | Compound Name | T. diversifolia | T. procumbens | A. robusta | Artificial Diet | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| D | F | C | D | F | C | D | F | C | D | F | C | ||||
| 1 | 5.7 | LC | quercetin 3-O-hexoside | ||||||||||||
| 2 | 7.7 | LC | flavonoid 3-O-methyl | ||||||||||||
| 3 | 7.9 | LC | flavonoid 3-O-methyl | ||||||||||||
| 4 | 8.2 | LC | luteolin | ||||||||||||
| 5 | 8.4 | LC | tagitinin A | ||||||||||||
| 6 | 8.5 | LC | nepetin | ||||||||||||
| 7 | 8.6 | LC | 5,7,3’,4’-tetrahydroxy 6,8-dimethoxyflavone | ||||||||||||
| 8 | 8.7 | LC | budlein A | ||||||||||||
| 9 | 8.7 | LC | tagitinin B | ||||||||||||
| 10 | 9.6 | LC | 1-hydroxy-3-O-methyltirotudin | ||||||||||||
| 11 | 9.8 | LC | hispidulin | ||||||||||||
| 12 | 10.0 | GC | 2,4-hexadienoic acid | ||||||||||||
| 13 | 10.2 | LC | acerosin | ||||||||||||
| 14 | 10.8 | LC | flavonoid 3-O-methyl | ||||||||||||
| 15 | 11.9 | GC | 2,6,6-trimethyl-2-cyclohexene-1,4-dione | ||||||||||||
| 16 | 12.6 | LC | nevadensin | ||||||||||||
| 17 | 12.6 | GC | borneol | ||||||||||||
| 18 | 13.2 | GC | catechol | ||||||||||||
| 19 | 13.4 | LC | kaur-15-ene 17,18 dioic acid | ||||||||||||
| 20 | 13.9 | LC | 16,17-dihydroxy-ent-kauran-19-oic acid | ||||||||||||
| 21 | 16.8 | GC | 2,4-decadienal (E,E) | ||||||||||||
| 22 | 18.9 | GC | ethanone,1-(1,6,7,7a-tetrahydro- 3,6,6-trimethylcyclopenta pyran-1-yl) | ||||||||||||
| 23 | 19.9 | LC | grandiflorenic acid | ||||||||||||
| 24 | 20.2 | GC | ethanone,1,1’-(1,4-phenylene) bis | ||||||||||||
| 25 | 20.4 | GC | benzoic acid, 4-hydroxy-methyl ester | ||||||||||||
| 26 | 21.0 | GC | germacrene D | ||||||||||||
| 27 | 21.0 | GC | 5,6-β-ionone epoxide | ||||||||||||
| 28 | 21.4 | GC | bicyclogermacrene | ||||||||||||
| 29 | 22.2 | GC | dihydroactinidiolide | ||||||||||||
| 30 | 23.4 | GC | spathulenol | ||||||||||||
| 31 | 25.2 | GC | α-cadinol | ||||||||||||
| 32 | 27.4 | GC | myristic acid | ||||||||||||
| 33 | 28.0 | GC | 2-cyclohexen-1-one, 4-hydroxy- 3,5,6-trimethyl-4-(3-oxo-1-butenyl) | ||||||||||||
| 34 | 29.1 | GC | neophytadiene | ||||||||||||
| 35 | 32.7 | GC | octadecanal | ||||||||||||
| 36 | 33.5 | GC | heptadecanoic acid | ||||||||||||
| 37 | 34.1 | GC | 9,12,15-octadecatrienoic acid, methyl ester | ||||||||||||
| 38 | 34.3 | GC | phytol | ||||||||||||
| 39 | 34.8 | GC | 9,12,15-octadecatrienoic acid | ||||||||||||
| 40 | 39.6 | GC | kaurenoic acid | ||||||||||||
| 41 | 40.6 | GC | 1-docosanol | ||||||||||||
| 8 | 45.8 | GC | budlein A | ||||||||||||
| 42 | 46.5 | GC | 1-hexacosanol | ||||||||||||
| 43 | 47.8 | GC | 1-heptacosanol | ||||||||||||
| 44 | 48.4 | GC | β-tocopherol | ||||||||||||
| 45 | 50.2 | GC | lathosterol | ||||||||||||
| 46 | 50.8 | GC | campesterol | ||||||||||||
| 47 | 51.9 | GC | chondrillasterol | ||||||||||||
| 48 | 52.1 | GC | β-amyrone | ||||||||||||
| 49 | 52.5 | GC | β-amyrin | ||||||||||||
| 50 | 52.8 | GC | α-amyrone | ||||||||||||
| 51 | 54.7 | GC | pseudotaraxasterol | ||||||||||||
| Diet | Mortality Rate (%) | Diapause Rate (%) | Larval Viability (%) a | Pupal Viability (%) b |
|---|---|---|---|---|
| T. diversifolia (1) | 0 | 0 | 100 | 100 |
| T. diversifolia (2) | 0 | 0 | 100 | 100 |
| T. diversifolia (3) | 0 | 5 | 95 | 100 |
| T. procumbens (1) | 25 | 0 | 75 | 100 |
| T. procumbens (2) | 30 | 10 | 60 | 100 |
| T. procumbens (3) | 5 | 15 | 80 | 100 |
| A. robusta (1) | 0 | 60 | 40 | 100 |
| A. robusta (2) | 0 | 75 | 25 | 100 |
| A. robusta (3) | 0 | 75 | 25 | 100 |
| Artificial diet (1) | 0 | 0 | 85 | 71 |
| Artificial diet (2) | 0 | 10 | 90 | 94 |
| Artificial diet (3) | 10 | 0 | 90 | 89 |
| Insect Stage | Caterpillars | Pupae | Adults | ||||
|---|---|---|---|---|---|---|---|
| 1st Instar | 2nd Instar | 3rd Instar | 4th Instar | 5th Instar | |||
| Weight (mg) | 1 to 10 | 10 to 20 | 20 to 50 | 50 to 120 | 120 to 200 | ||
| Duration (days) | |||||||
| T. diversifolia | 5.85 ± 0.67 | 3.70 ± 0.66 | 3.25 ± 0.44 | 3.30 ± 0.66 | 3.35 ± 0.49 | 7.55 ± 0.60 | 11.35 ± 1.04 |
| T. procumbens | 12.25 ± 0.79 | 2.70 ± 0.66 | 3.30 ± 0.47 | 3.20 ± 0.62 | 3.05 ± 0.51 | 7.45 ± 0.60 | 11.70 ± 0.73 |
| A. robusta | 15.20 ± 0.41 | 4.10 ± 0.55 | 4.15 ± 0.59 | 4.10 ± 0.64 | 2.65 ± 0.67 | 7.70 ± 0.73 | 11.75 ± 0.85 |
| Artificial diet | 12.10 ± 0.72 | 2.60 ± 0.68 | 3.35 ± 0.49 | 3.25 ± 0.44 | 2.65 ± 0.49 | 7.90 ± 0.55 | 11.65 ± 0.99 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gallon, M.E.; Gobbo-Neto, L. Plant Metabolites Involved in the Differential Development of a Heliantheae-Specialist Insect. Metabolites 2021, 11, 134. https://doi.org/10.3390/metabo11030134
Gallon ME, Gobbo-Neto L. Plant Metabolites Involved in the Differential Development of a Heliantheae-Specialist Insect. Metabolites. 2021; 11(3):134. https://doi.org/10.3390/metabo11030134
Chicago/Turabian StyleGallon, Marília Elias, and Leonardo Gobbo-Neto. 2021. "Plant Metabolites Involved in the Differential Development of a Heliantheae-Specialist Insect" Metabolites 11, no. 3: 134. https://doi.org/10.3390/metabo11030134
APA StyleGallon, M. E., & Gobbo-Neto, L. (2021). Plant Metabolites Involved in the Differential Development of a Heliantheae-Specialist Insect. Metabolites, 11(3), 134. https://doi.org/10.3390/metabo11030134