Inhibition of Ganglioside Synthesis Suppressed Liver Cancer Cell Proliferation through Targeting Kinetochore Metaphase Signaling
Abstract
:1. Introduction
2. Results
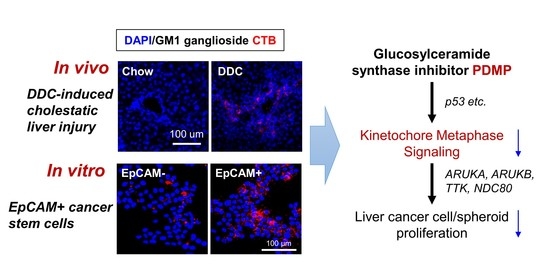
2.1. Hepatic Ganglioside Synthesis Was Increased along with the Expansion of Mouse Hepatic Stem/Progenitor Cells
2.2. Ganglioside Synthesis Was Increased in Human Liver Cancer Stem-Like Cells
2.3. Inhibition of Ganglioside Synthesis Suppressed the Proliferation of Liver Cancer Cells
2.4. Inhibition of Ganglioside Synthesis Affected the Lipid Composition of Liver Cancer Cells
2.5. Proteome-Based Analysis of Downstream Signaling Pathway Following Ganglioside Synthesis Inhibition in Liver Cancer Cells
2.6. Inhibition of Ganglioside Synthesis Targets Chromosome Segregation Signaling Pathway in Liver Cancer Cells
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Animal Experiments
4.3. Serum ALT Activity
4.4. RNA Isolation and Real-Time PCR
4.5. Immunofluorescence Staining
4.6. Cell Culture
4.7. Cell Sorting
4.8. 3D Spheroid Cultures
4.9. Spheroid Proliferation Assay
4.10. Annexin V Staining Assay
4.11. Lipid Extraction
4.12. MALDI-TOFMS-Based Lipidomic Analysis
4.13. nLC-MS/MS-Based Proteomic Analysis
4.14. Knowledge-Based Pathway Analysis
4.15. Western Bloting
4.16. Multivariate and Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Venook, A.P.; Papandreou, C.; Furuse, J.; Ladrón de Guevara, L. The Incidence and Epidemiology of Hepatocellular Carcinoma: A Global and Regional Perspective. Oncologist 2010, 15, 5–13. [Google Scholar] [CrossRef]
- Llovet, J.M.; Villanueva, A.; Lachenmayer, A.; Finn, R.S. Advances in targeted therapies for hepatocellular carcinoma in the genomic era. Nat. Rev. Clin. Oncol. 2015, 12, 408–424. [Google Scholar] [CrossRef] [PubMed]
- Hashim, D.; Boffetta, P.; La Vecchia, C.; Rota, M.; Bertuccio, P.; Malvezzi, M.; Negri, E. The global decrease in cancer mortality: Trends and disparities. Ann. Oncol. 2016, 27, 926–933. [Google Scholar] [CrossRef] [PubMed]
- Starley, B.Q.; Calcagno, C.J.; Harrison, S.A. Nonalcoholic fatty liver disease and hepatocellular carcinoma: A weighty connection. Hepatology 2010, 51, 1820–1832. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.Y.; Tatsukawa, H.; Hitomi, K.; Shirakami, Y.; Ishibashi, N.; Shimizu, M.; Moriwaki, H.; Kojima, S. Metabolome Analyses Uncovered a Novel Inhibitory Effect of Acyclic Retinoid on Aberrant Lipogenesis in a Mouse Diethylnitrosamine-Induced Hepatic Tumorigenesis Model. Cancer Prev. Res. 2016, 9, 205–214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carroll, P.A.; Diolaiti, D.; McFerrin, L.; Gu, H.; Djukovic, D.; Du, J.; Cheng, P.F.; Anderson, S.; Ulrich, M.; Hurley, J.B.; et al. Deregulated Myc Requires MondoA/Mlx for Metabolic Reprogramming and Tumorigenesis. Cancer Cell 2015, 27, 271–285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lai, K.K.Y.; Kweon, S.-M.; Chi, F.; Hwang, E.; Kabe, Y.; Higashiyama, R.; Qin, L.; Yan, R.; Wu, R.P.; Lai, K.; et al. Stearoyl-CoA Desaturase Promotes Liver Fibrosis and Tumor Development in Mice via a Wnt Positive-Signaling Loop by Stabilization of Low-Density Lipoprotein-Receptor-Related Proteins 5 and 6. Gastroenterology 2017, 152, 1477–1491. [Google Scholar] [CrossRef] [Green Version]
- Van Meer, G.; Voelker, D.R.; Feigenson, G.W. Membrane lipids: Where they are and how they behave. Nat. Rev. Mol. Cell Biol. 2008, 9, 112–124. [Google Scholar] [CrossRef] [PubMed]
- Tummala, K.S.; Brandt, M.; Teijeiro, A.; Graña, O.; Schwabe, R.F.; Perna, C.; Djouder, N. Hepatocellular Carcinomas Originate Predominantly from Hepatocytes and Benign Lesions from Hepatic Progenitor Cells. Cell Rep. 2017, 19, 584–600. [Google Scholar] [CrossRef] [Green Version]
- Nakagawa, H.; Hayata, Y.; Kawamura, S.; Yamada, T.; Fujiwara, N.; Koike, K. Lipid Metabolic Reprogramming in Hepatocellular Carcinoma. Cancers 2018, 10, 447. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sonnino, S.; Prinetti, A. Gangliosides as Regulators of Cell Membrane Organization and Functions. In Sphingolipids as Signaling and Regulatory Molecules; Springer: Berlin/Heidelberg, Germany, 2010; Volume 688, pp. 165–184. [Google Scholar]
- Julien, S.; Bobowski, M.; Steenackers, A.; Le Bourhis, X.; Delannoy, P. How Do Gangliosides Regulate RTKs Signaling? Cells 2013, 2, 751–767. [Google Scholar] [CrossRef] [Green Version]
- Simons, K.; Toomre, D. Lipid rafts and signal transduction. Nat. Rev. Mol. Cell Biol. 2000, 1, 31–39. [Google Scholar] [CrossRef]
- Kwak, D.H.; Seo, B.B.; Chang, K.T.; Choo, Y.K. Roles of gangliosides in mouse embryogenesis and embryonic stem cell differentiation. Exp. Mol. Med. 2011, 43, 379. [Google Scholar] [CrossRef] [Green Version]
- Schengrund, C.-L. Gangliosides: Glycosphingolipids essential for normal neural development and function. Trends Biochem. Sci. 2015, 40, 397–406. [Google Scholar] [CrossRef]
- Hermetet, F.; Buffière, A.; Aznague, A.; Pais de Barros, J.-P.; Bastie, J.-N.; Delva, L.; Quéré, R. High-fat diet disturbs lipid raft/TGF-β signaling-mediated maintenance of hematopoietic stem cells in mouse bone marrow. Nat. Commun. 2019, 10, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Yamazaki, S.; Iwama, A.; Takayanagi, S.-I.; Morita, Y.; Eto, K.; Ema, H.; Nakauchi, H. Cytokine signals modulated via lipid rafts mimic niche signals and induce hibernation in hematopoietic stem cells. EMBO J. 2006, 25, 3515–3523. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Groux-Degroote, S.; Rodríguez-Walker, M.; Dewald, J.H.; Daniotti, J.L.; Delannoy, P. Gangliosides in Cancer Cell Signaling. Prog. Mol. Biol. Transl. Sci. 2018, 156, 197–227. [Google Scholar] [CrossRef]
- Cavdarli, S.; Delannoy, P.; Groux-Degroote, S. O-acetylated Gangliosides as Targets for Cancer Immunotherapy. Cells 2020, 9, 741. [Google Scholar] [CrossRef] [Green Version]
- Jennemann, R.; Federico, G.; Mathow, D.; Rabionet, M.; Rampoldi, F.; Popovic, Z.V.; Volz, M.; Hielscher, T.; Sandhoff, R.; Gröne, H.-J. Inhibition of hepatocellular carcinoma growth by blockade of glycosphingolipid synthesis. Oncotarget 2017, 8, 109201–109216. [Google Scholar] [CrossRef] [Green Version]
- Deng, X.; Zhang, X.; Li, W.; Feng, R.-X.; Li, L.; Yi, G.-R.; Zhang, X.-N.; Yin, C.; Yu, H.-Y.; Zhang, J.-P.; et al. Chronic Liver Injury Induces Conversion of Biliary Epithelial Cells into Hepatocytes. Cell Stem Cell 2018, 23, 114–122. [Google Scholar] [CrossRef] [Green Version]
- Sato, K.; Marzioni, M.; Meng, F.; Francis, H.; Glaser, S.; Alpini, G. Ductular Reaction in Liver Diseases: Pathological Mechanisms and Translational Significances. Hepatology 2019, 69, 420–430. [Google Scholar] [CrossRef] [Green Version]
- Luedde, T.; Kaplowitz, N.; Schwabe, R.F. Cell Death and Cell Death Responses in Liver Disease: Mechanisms and Clinical Relevance. Gastroenterology 2014, 147, 765–783. [Google Scholar] [CrossRef] [Green Version]
- Chiricozzi, E.; Lunghi, G.; Di Biase, E.; Fazzari, M.; Sonnino, S.; Mauri, L. GM1 Ganglioside Is A Key Factor in Maintaining the Mammalian Neuronal Functions Avoiding Neurodegeneration. Int. J. Mol. Sci. 2020, 21, 868. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verma, M.; Schneider, J.S. siRNA-mediated knockdown of B3GALT4 decreases GM1 ganglioside expression and enhances vulnerability for neurodegeneration. Mol. Cell. Neurosci. 2019, 95, 25–30. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, Y.; Ninomiya, H.; Ohsaki, Y.; Higaki, K.; Davies, J.P.; Ioannou, Y.A.; Ohno, K. Accumulation of cholera toxin and GM1 ganglioside in the early endosome of Niemann-Pick C1-deficient cells. Proc. Natl. Acad. Sci. USA 2001, 98, 12391–12396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boulter, L.; Govaere, O.; Bird, T.G.; Radulescu, S.; Ramachandran, P.; Pellicoro, A.; Ridgway, R.A.; Seo, S.S.; Spee, B.; Van Rooijen, N.; et al. Macrophage-derived Wnt opposes Notch signaling to specify hepatic progenitor cell fate in chronic liver disease. Nat. Med. 2012, 18, 572–579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hernandez, C.; Huebener, P.; Pradere, J.-P.; Antoine, D.J.; Friedman, R.A.; Schwabe, R.F. HMGB1 links chronic liver injury to progenitor responses and hepatocarcinogenesis. J. Clin. Investig. 2018, 128, 2436–2451. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, K.; Suzuki, A.M.; Dos Santos, A.; Desterke, C.; Collino, A.; Ghisletti, S.; Braun, E.; Bonetti, A.; Fort, A.; Qin, X.Y.; et al. CAGE profiling of ncRNAs in hepatocellular carcinoma reveals widespread activation of retroviral LTR promoters in virus-induced tumors. Genome Res. 2015, 25, 1812–1824. [Google Scholar] [CrossRef] [Green Version]
- Qin, X.Y.; Suzuki, H.; Honda, M.; Okada, H.; Kaneko, S.; Inoue, I.; Ebisui, E.; Hashimoto, K.; Carninci, P.; Kanki, K.; et al. Prevention of hepatocellular carcinoma by targeting MYCN-positive liver cancer stem cells with acyclic retinoid. Proc. Natl. Acad. Sci. USA 2018, 115, 4969–4974. [Google Scholar] [CrossRef] [Green Version]
- Yamashita, T.; Ji, J.; Budhu, A.; Forgues, M.; Yang, W.; Wang, H.Y.; Jia, H.; Ye, Q.; Qin, L.X.; Wauthier, E.; et al. EpCAM-Positive Hepatocellular Carcinoma Cells Are Tumor-Initiating Cells with Stem/Progenitor Cell Features. Gastroenterology 2009, 136, 1012–1024. [Google Scholar] [CrossRef] [Green Version]
- Qin, X.-Y.; Su, T.; Yu, W.; Kojima, S. Lipid desaturation-associated endoplasmic reticulum stress regulates MYCN gene expression in hepatocellular carcinoma cells. Cell Death Dis. 2020, 11, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Qin, X.-Y.; Kojima, S. Inhibition of Stearoyl-CoA Desaturase-1 Activity Suppressed SREBP Signaling in Colon Cancer Cells and Their Spheroid Growth. Gastrointest. Disord. 2019, 1, 14. [Google Scholar] [CrossRef] [Green Version]
- Fuchs, B.; Schiller, J. MALDI-TOF MS Analysis of Lipids from Cells, Tissues and Body Fluids. Lipids Health Dis. 2008, 49, 541–565. [Google Scholar] [CrossRef]
- Krämer, A.; Green, J.; Pollard, J.; Tugendreich, S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef]
- Kikegawa, M.; Qin, X.-Y.; Ito, T.; Nishikawa, H.; Nansai, H.; Sone, H. Early Transcriptomic Changes upon Thalidomide Exposure Influence the Later Neuronal Development in Human Embryonic Stem Cell-Derived Spheres. Int. J. Mol. Sci. 2020, 21, 5564. [Google Scholar] [CrossRef]
- Giannakakou, P.; Nakano, M.; Nicolaou, K.C.; O’Brate, A.; Yu, J.; Blagosklonny, M.V.; Greber, U.F.; Fojo, T. Enhanced microtubule-dependent trafficking and p53 nuclear accumulation by suppression of microtubule dynamics. Proc. Natl. Acad. Sci. USA 2002, 99, 10855–10860. [Google Scholar] [CrossRef] [Green Version]
- Maiato, H. The dynamic kinetochore-microtubule interface. J. Cell Sci. 2004, 117, 5461–5477. [Google Scholar] [CrossRef] [Green Version]
- Yuen, K.W.Y.; Montpetit, B.; Hieter, P. The kinetochore and cancer: What’s the connection? Curr. Opin. Cell Biol. 2005, 17, 576–582. [Google Scholar] [CrossRef]
- Cimini, D. Merotelic kinetochore orientation, aneuploidy, and cancer. Biochim. Biophys. Acta Rev. Cancer 2008, 1786, 32–40. [Google Scholar] [CrossRef]
- Zhang, W.; Mao, J.-H.; Zhu, W.; Jain, A.K.; Liu, K.; Brown, J.B.; Karpen, G.H. Centromere and kinetochore gene misexpression predicts cancer patient survival and response to radiotherapy and chemotherapy. Nat. Commun. 2016, 7, 1–15. [Google Scholar] [CrossRef]
- Oser, M.G.; Fonseca, R.; Chakraborty, A.A.; Brough, R.; Spektor, A.; Jennings, R.B.; Flaifel, A.; Novak, J.S.; Gulati, A.; Buss, E.; et al. Cells Lacking the RB1 Tumor Suppressor Gene Are Hyperdependent on Aurora B Kinase for Survival. Cancer Discov. 2019, 9, 230–247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maia, A.R.R.; de Man, J.; Boon, U.; Janssen, A.; Song, J.Y.; Omerzu, M.; Sterrenburg, J.G.; Prinsen, M.B.W.; Willemsen-Seegers, N.; de Roos, J.A.D.M.; et al. Inhibition of the spindle assembly checkpoint kinase TTK enhances the efficacy of docetaxel in a triple-negative breast cancer model. Ann. Oncol. 2015, 26, 2180–2192. [Google Scholar] [CrossRef] [PubMed]
- Chuang, S.-H.; Lee, Y.-S.E.; Huang, L.Y.L.; Chen, C.-K.; Lai, C.-L.; Lin, Y.-H.; Yang, J.-Y.; Yang, S.-C.; Chang, L.-H.; Chen, C.-H.; et al. Discovery of T-1101 tosylate as a first-in-class clinical candidate for Hec1/Nek2 inhibition in cancer therapy. Eur. J. Med. Chem. 2020, 191, 112118. [Google Scholar] [CrossRef] [PubMed]
- Gustafson, W.C.; Meyerowitz, J.G.; Nekritz, E.A.; Chen, J.; Benes, C.; Charron, E.; Simonds, E.F.; Seeger, R.; Matthay, K.K.; Hertz, N.T.; et al. Drugging MYCN through an Allosteric Transition in Aurora Kinase. Cancer Cell 2014, 26, 414–427. [Google Scholar] [CrossRef] [Green Version]
- Jiang, J.; Wang, J.; Yue, M.; Cai, X.; Wang, T.; Wu, C.; Su, H.; Wang, Y.; Han, M.; Zhang, Y.; et al. Direct Phosphorylation and Stabilization of MYC by Aurora B Kinase Promote T-cell Leukemogenesis. Cancer Cell 2020, 37, 200–215.e205. [Google Scholar] [CrossRef]
- Horwacik, I.; Durbas, M.; Boratyn, E.; Węgrzyn, P.; Rokita, H. Targeting GD2 ganglioside and aurora A kinase as a dual strategy leading to cell death in cultures of human neuroblastoma cells. Cancer Lett. 2013, 341, 248–264. [Google Scholar] [CrossRef] [PubMed]
- Spurgers, K.B.; Gold, D.L.; Coombes, K.R.; Bohnenstiehl, N.L.; Mullins, B.; Meyn, R.E.; Logothetis, C.J.; McDonnell, T.J. Identification of Cell Cycle Regulatory Genes as Principal Targets of p53-mediated Transcriptional Repression. J. Biol. Chem. 2006, 281, 25134–25142. [Google Scholar] [CrossRef] [Green Version]
- Rivlin, N.; Brosh, R.; Oren, M.; Rotter, V. Mutations in the p53 Tumor Suppressor Gene: Important Milestones at the Various Steps of Tumorigenesis. Genes Cancer 2011, 2, 466–474. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, K.; Yan, Y.; Yuan, B.; Dasgupta, A.; Sun, J.; Mu, H.; Do, K.-A.; Ueno, N.T.; Andreeff, M.; Battula, V.L. ST8SIA1 Regulates Tumor Growth and Metastasis in TNBC by Activating the FAK–AKT–mTOR Signaling Pathway. Mol. Cancer Ther. 2018, 17, 2689–2701. [Google Scholar] [CrossRef] [Green Version]
- Yu, A.L.; Gilman, A.L.; Ozkaynak, M.F.; London, W.B.; Kreissman, S.G.; Chen, H.X.; Smith, M.; Anderson, B.; Villablanca, J.G.; Matthay, K.K.; et al. Anti-GD2 Antibody with GM-CSF, Interleukin-2, and Isotretinoin for Neuroblastoma. N. Engl. J. Med. 2010, 363, 1324–1334. [Google Scholar] [CrossRef] [Green Version]
- Takehara, T.; Tatsumi, T.; Suzuki, T.; Rucker, E.B.; Hennighausen, L.; Jinushi, M.; Miyagi, T.; Kanazawa, Y.; Hayashi, N. Hepatocyte-specific disruption of Bcl-xL leads to continuous hepatocyte apoptosis and liver fibrotic responses. Gastroenterology 2004, 127, 1189–1197. [Google Scholar] [CrossRef]
- Pusterla, T.; Nèmeth, J.; Stein, I.; Wiechert, L.; Knigin, D.; Marhenke, S.; Longerich, T.; Kumar, V.; Arnold, B.; Vogel, A.; et al. Receptor for advanced glycation endproducts (RAGE) is a key regulator of oval cell activation and inflammation-associated liver carcinogenesis in mice. Hepatology 2013, 58, 363–373. [Google Scholar] [CrossRef]
- Krysko, O.; Løve Aaes, T.; Bachert, C.; Vandenabeele, P.; Krysko, D.V. Many faces of DAMPs in cancer therapy. Cell Death Dis. 2013, 4, e631. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koberlin, M.S.; Snijder, B.; Heinz, L.X.; Baumann, C.L.; Fauster, A.; Vladimer, G.I.; Gavin, A.C.; Superti-Furga, G. A Conserved Circular Network of Coregulated Lipids Modulates Innate Immune Responses. Cell 2015, 162, 170–183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gebai, A.; Gorelik, A.; Li, Z.; Illes, K.; Nagar, B. Structural basis for the activation of acid ceramidase. Nat. Commun. 2018, 9, 1621. [Google Scholar] [CrossRef]
- Bedia, C.; Casas, J.; Andrieu-Abadie, N.; Fabrias, G.; Levade, T. Acid ceramidase expression modulates the sensitivity of A375 melanoma cells to dacarbazine. J. Biol. Chem. 2011, 286, 28200–28209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okino, N.; He, X.; Gatt, S.; Sandhoff, K.; Ito, M.; Schuchman, E.H. The reverse activity of human acid ceramidase. J. Biol. Chem. 2003, 278, 29948–29953. [Google Scholar] [CrossRef] [Green Version]
- Llacuna, L.; Mari, M.; Garcia-Ruiz, C.; Fernandez-Checa, J.C.; Morales, A. Critical role of acidic sphingomyelinase in murine hepatic ischemia-reperfusion injury. Hepatology 2006, 44, 561–572. [Google Scholar] [CrossRef] [Green Version]
- Mayo, L.; Trauger, S.A.; Blain, M.; Nadeau, M.; Patel, B.; Alvarez, J.I.; Mascanfroni, I.D.; Yeste, A.; Kivisäkk, P.; Kallas, K.; et al. Regulation of astrocyte activation by glycolipids drives chronic CNS inflammation. Nat. Med. 2014, 20, 1147–1156. [Google Scholar] [CrossRef] [Green Version]
- Plo, I.; Lehne, G.; Beckstrom, K.J.; Maestre, N.; Bettaieb, A.; Laurent, G.; Lautier, D. Influence of ceramide metabolism on P-glycoprotein function in immature acute myeloid leukemia KG1a cells. Mol. Pharmacol. 2002, 62, 304–312. [Google Scholar] [CrossRef] [Green Version]
- Nagafuku, M.; Kabayama, K.; Oka, D.; Kato, A.; Tanichi, S.; Shimada, Y.; Ohno-Iwashita, Y.; Yamasaki, S.; Saito, T.; Iwabuchi, K.; et al. Reduction of glycosphingolipid levels in lipid rafts affects the expression state and function of glycosylphosphatidylinositol-anchored proteins but does not impair signal transduction via the T cell receptor. J. Biol. Chem. 2003, 278, 51920–51927. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawashima, N.; Nishimiya, Y.; Takahata, S.; Nakayama, K.I. Induction of Glycosphingolipid GM3 Expression by Valproic Acid Suppresses Cancer Cell Growth. J. Biol. Chem. 2016, 291, 21424–21433. [Google Scholar] [CrossRef] [Green Version]
- Fedoryszak-Kuska, N.; Panasiewicz, M.; Domek, H.; Pacuszka, T. Glucosylceramide synthase inhibitors D-PDMP and D-EtDO-P4 decrease the GM3 ganglioside level, differ in their effects on insulin receptor autophosphorylation but increase Akt1 kinase phosphorylation in human hepatoma HepG2 cells. Acta Biochim. Pol. 2016, 63, 247–251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gong, L.; Yang, B.; Xu, M.; Cheng, B.; Tang, X.; Zheng, P.; Jing, Y.; Wu, G.J. Bortezomib-induced apoptosis in cultured pancreatic cancer cells is associated with ceramide production. Cancer Chemother. Pharmacol. 2014, 73, 69–77. [Google Scholar] [CrossRef]
- Griner, R.D.; Bollag, W.B. Inhibition of [(3)H]thymidine transport is a nonspecific effect of PDMP in primary cultures of mouse epidermal keratinocytes. J. Pharmacol. Exp. Ther. 2000, 294, 1219–1224. [Google Scholar] [PubMed]
- Chatterjee, S.; Alsaeedi, N.; Hou, J.; Bandaru, V.V.; Wu, L.; Halushka, M.K.; Pili, R.; Ndikuyeze, G.; Haughey, N.J. Use of a glycolipid inhibitor to ameliorate renal cancer in a mouse model. PLoS ONE 2013, 8, e63726. [Google Scholar] [CrossRef] [Green Version]
- Bedja, D.; Yan, W.; Lad, V.; Iocco, D.; Sivakumar, N.; Bandaru, V.V.R.; Chatterjee, S. Inhibition of glycosphingolipid synthesis reverses skin inflammation and hair loss in ApoE-/-mice fed western diet. Sci. Rep. 2018, 8, 11463. [Google Scholar] [CrossRef]
- Le Stunff, H.; Galve-Roperh, I.; Peterson, C.; Milstien, S.; Spiegel, S. Sphingosine-1-phosphate phosphohydrolase in regulation of sphingolipid metabolism and apoptosis. J. Cell. Biol. 2002, 158, 1039–1049. [Google Scholar] [CrossRef] [PubMed]
- Simon, J.; Ouro, A.; Ala-Ibanibo, L.; Presa, N.; Delgado, T.C.; Martinez-Chantar, M.L. Sphingolipids in Non-Alcoholic Fatty Liver Disease and Hepatocellular Carcinoma: Ceramide Turnover. Int. J. Mol. Sci. 2019, 21, 40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, L.; Jin, G.H.; Zhou, J.Y. The Role of Ceramide in the Pathogenesis of Alcoholic Liver Disease. Alcohol Alcohol. 2016, 51, 251–257. [Google Scholar] [CrossRef] [Green Version]
- Ismail, I.T.; Elfert, A.; Helal, M.; Salama, I.; El-Said, H.; Fiehn, O. Remodeling Lipids in the Transition from Chronic Liver Disease to Hepatocellular Carcinoma. Cancers 2020, 13, 88. [Google Scholar] [CrossRef] [PubMed]
- Fujise, K.; Nagamori, S.; Hasumura, S.; Homma, S.; Sujino, H.; Matsuura, T.; Shimizu, K.; Niiya, M.; Kameda, H.; Fujita, K.; et al. Integration of hepatitis B virus DNA into cells of six established human hepatocellular carcinoma cell lines. Hepatogastroenterology 1990, 37, 457–460. [Google Scholar]
- Gil, M.; Samino, S.; Barrilero, R.; Correig, X. Lipid Profiling Using 1H NMR Spectroscopy. Methods Mol. Biol. 2019, 2037, 35–47. [Google Scholar] [CrossRef]
- Wiśniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.-Y.; Hara, M.; Arner, E.; Kawaguchi, Y.; Inoue, I.; Tatsukawa, H.; Furutani, Y.; Nagatsuma, K.; Matsuura, T.; Wei, F.; et al. Transcriptome Analysis Uncovers a Growth-Promoting Activity of Orosomucoid-1 on Hepatocytes. EBioMedicine 2017, 24, 257–266. [Google Scholar] [CrossRef] [PubMed] [Green Version]





| Gene Symbol | Forward | Reverse |
|---|---|---|
| Mouse | ||
| 16s | AGGAGCGATTTGCTGGTGTGGA | GCTACCAGGGCCTTTGAGATGGA |
| Mki67 | AGGGTAACTCGTGGAACCAA | TTAACTTCTTGGTGCATACAATGTC |
| Adgre1 | TGTCTGACAATTGGGATCTGCCCT | ATAGCTTCCGAGAGTGTTGTGGCA |
| Tnfα | TGTCTACTCCCAGGTTCTCT | GGGGCAGGGGCTCTTGAC |
| Acta2 | CGATAGAACACGGCATCATC | CATCAGGCAGTTCGTAGCTC |
| Mmp9 | CCCATGTCACTTTCCCTTCAC | GCCGTCCTTATCGTAGTCAGC |
| Cers1 | CCACCACACACATCTTTCGG | GGAGCAGGTAAGCGCAGTAG |
| Cers2 | ATGCTCCAGACCTTGTATGACT | CTGAGGCTTTGGCATAGACAC |
| Cers3 | ATGGGCTTGTCTTCGTGAAAG | TTGCTTGTGGAATGCTTGAAAAA |
| Cers4 | TACCCACATCAGACCCTGAAT | TGAAGTCCTTGCGTTTGACATC |
| Cers5 | CGGGGAAAGGTGTCTAAGGAT | GTTCATGCAGTTGGCACCATT |
| Asah1 | CGTGGACAGAAGATTGCAGAA | TGGTGCCTTTTGAGCCAATAAT |
| Naaa | GACTCCGCCTCTCTTCAACG | ACCATCCCGAGTACCCACTG |
| B3galt4 | GGCAGTGCCCCTTCTGTATTT | CGAGGCATAGGGTGGAAAAG |
| St3gal2 | CACCCTGACTCGGCTGCTT | TCTCGCGCCTTAGGGCTAA |
| Human | ||
| ACTB | GCACAGAGCCTCGCCTT | GTTGTCGACGACGAGCG |
| AURKA | GCCCTGTCTTACTGTCATTCG | AGAGAGTGGTCCTCCTGGAAG |
| AURKB | ATCTGCTCTTAGGGCCAAGGG | CACATTGTCTTCCTCCTCAGGG |
| NCD80 | TCAAGGACCCGAGACCACTTA | GGGAGCTTGTAGAGATTTCATGG |
| TTK | TGGCCAACCTGCCTGTTT | AATGCATTCATTTGCTGAAGAAGA |
| TP53 | GGCCCACTTCACCGTACTAA | GTGGTTTCAAGGCCAGATGT |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Su, T.; Qin, X.-Y.; Dohmae, N.; Wei, F.; Furutani, Y.; Kojima, S.; Yu, W. Inhibition of Ganglioside Synthesis Suppressed Liver Cancer Cell Proliferation through Targeting Kinetochore Metaphase Signaling. Metabolites 2021, 11, 167. https://doi.org/10.3390/metabo11030167
Su T, Qin X-Y, Dohmae N, Wei F, Furutani Y, Kojima S, Yu W. Inhibition of Ganglioside Synthesis Suppressed Liver Cancer Cell Proliferation through Targeting Kinetochore Metaphase Signaling. Metabolites. 2021; 11(3):167. https://doi.org/10.3390/metabo11030167
Chicago/Turabian StyleSu, Ting, Xian-Yang Qin, Naoshi Dohmae, Feifei Wei, Yutaka Furutani, Soichi Kojima, and Wenkui Yu. 2021. "Inhibition of Ganglioside Synthesis Suppressed Liver Cancer Cell Proliferation through Targeting Kinetochore Metaphase Signaling" Metabolites 11, no. 3: 167. https://doi.org/10.3390/metabo11030167
APA StyleSu, T., Qin, X. -Y., Dohmae, N., Wei, F., Furutani, Y., Kojima, S., & Yu, W. (2021). Inhibition of Ganglioside Synthesis Suppressed Liver Cancer Cell Proliferation through Targeting Kinetochore Metaphase Signaling. Metabolites, 11(3), 167. https://doi.org/10.3390/metabo11030167