HPTLC-Based Chemical Profiling: An Approach to Monitor Plant Metabolic Expansion Caused by Fungal Endophytes
Abstract
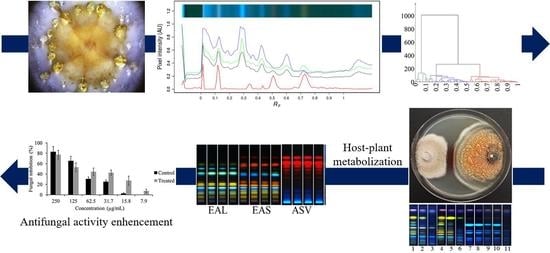
:1. Introduction
2. Results
2.1. Endophyte Isolation and HPTLC Multivariate Data Analysis
2.2. HPTLC-Fungal Growth Correlation and Co-culture Bioassays
2.3. Fungal Feeding Experiments and Antimicrobial Activity
3. Discussion
4. Materials and Methods
4.1. Fungal Endophyte Isolation
4.2. Endophytes-Fungal Pathogens Co-culture Assays
4.3. Plant Extracts Feeding Experiments
4.4. 1H NMR Analysis
4.5. High Performance Thin Layer Chromatography
4.6. Microorganisms
4.7. Antibacterial Activity
4.8. Antifungal Activity and Minimum Effective Concentration (MEC)
4.8.1. Spore Production
4.8.2. Minimum Effective Concentration (MEC)
4.8.3. Fungi Identification
4.9. Data Processing and Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gholson, J.L.; Muir, T.W. Chemical signaling among bacteria and its inhibition. Chem. Biol. 2003, 10, 1007–1021. [Google Scholar]
- Kingston, D.G.I. Modern natural products drug discovery and its relevance to biodiversity conservation. J. Nat. Prod. 2011, 74, 496–511. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, C.; Zacchetti, B.; Ram, A.F.J.; van Wezel, G.P.; Claessen, D.; Choi, Y.H. Expanding the chemical space for natural products by Aspergillus–Streptomyces co–cultivation and biotransformation. Sci. Rep. 2015, 5, 1086. [Google Scholar] [CrossRef] [Green Version]
- Grigoriev, I. Fungal genomics for energy and environment. In Genomics of Soil– and Plant–Associated Fungi: Soil Biology, 36th ed.; Horwitz, B., Mukherjee, P., Mukherjee, M., Eds.; Springer: Berlin/Heidelberg, Germany, 2013; pp. 11–27. [Google Scholar]
- Zeilinger, S.; Gupta, V.K.; Dahms, T.E.S.; Silva, R.N.; Singh, H.B.; Upadhyay, R.S.; Vieira–Gomes, E.; Tsui, C.K.M.; Nayak, C. Friends or foes? Emerging insights from fungal interactions with plants. FEMS Microbiol. Rev. 2016, 40, 182–207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilson, D. Endophyte—The evolution of the term; a clarification of its use and definition. Oikos 1995, 73, 274–276. [Google Scholar] [CrossRef]
- Das, A.; Varma, A. Symbiosis: The art of living. In Symbiotic Fungi Principles and Practice, 18th ed.; Varma, A., Kharkwal, A.C., Eds.; Springer: Berlin, Germany, 2009; pp. 1–28. [Google Scholar]
- Kusari, S.; Zühlke, S.; Spiteller, M. An endophytic fungus from Camptotheca acuminata that produces camptothecin and analogues. J. Nat. Prod. 2009, 72, 2–7. [Google Scholar] [CrossRef]
- Eyberger, A.L.; Dondapati, R.; Porte, J.R. Endophyte fungal isolates from Podophyllum peltatum produce podophyllotoxin. J. Nat. Prod. 2006, 69, 1121–1124. [Google Scholar] [CrossRef]
- Stierle, A.; Strobel, G.; Stierle, D. Taxol and taxane production by Taxomyces andreanae an endophytic fungus of Pacific yew. Science 1993, 260, 214–216. [Google Scholar] [CrossRef]
- Pieterse, C.M.J.; Zamioudis, C.; Berendsen, R.L.; Weller, D.M.; Van Wees, S.C.M.; Bakker, P.A.H.M. Induced systemic resistance by beneficial microbes. Annu. Rev. Phytopathol. 2014, 52, 347–375. [Google Scholar] [CrossRef] [Green Version]
- Kucht, S.; Gross, J.; Hussein, Y.; Grothe, T.; Keller, U.; Basar, S.; König, W.A.; Steiner, U.; Leistner, E. Elimination of ergoline alkaloids following treatment of Ipomoea asarifolia (Convolvulaceae) with fungicides. Planta 2004, 219, 619–625. [Google Scholar] [CrossRef]
- Salomé–Abarca, L.F.; van der Pas, J.; Kim, H.K.; van Uffelen, G.A.; Klinkhamer, P.G.L.; Choi, Y.H. Metabolic discrimination of pine resins using multiple analytical platforms. Phytochemistry 2018, 155, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Gromski, P.S.; Muhamadali, H.; Ellis, D.I.; Xu, Y.; Correa, E.; Turner, M.L.; Goodacre, R. A tutorial review: Metabolomics and partial least squares–discriminant analysis a marriage of convenience or a shotgun wedding. Anal Chim Acta. 2015, 16, 10–23. [Google Scholar] [CrossRef]
- Ge, Y.; Sun, M.; Salomé–Abarca, L.F.; Wang, M.; Choi, Y.H. Investigation of species and environmental effects on rhubarb roots metabolome using 1H NMR combined with high performance thin layer chromatography. Metabolomics 2018, 14, 137. [Google Scholar] [CrossRef] [Green Version]
- Dey, A.; Hazrab, A.K.; Nandya, S.; Kaur, P.; Pandey, D.K. Selection of elite germplasms for industrially viable medicinal crop Bacopa monnieri for bacoside A production: An HPTLC–coupled chemotaxonomic study. Ind. Crops Prod. 2020, 158, e112975. [Google Scholar] [CrossRef]
- Mulaudzi, N.; Anokwuru, C.P.; Tankeu, S.Y.; Combrinck, S.; Chen, W.; Vermaak, I.; Viljoen, A.M. Phytochemical Profiling and Quality Control of Terminalia sericea Burch. ex DC. Using HPTLC Metabolomics. Molecules 2021, 26, 432. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Ahlgren, S.; Korthout, H.A.A.J.; Salomé–Abarca, L.F.; Bayona, L.M.; Verpoorte, R.; Choi, Y.H. Broad range chemical profiling of natural deep eutectic solvent extracts using a high performance thin layer chromatography–based method. J. Chromatogr. A 2018, 1532, 198–207. [Google Scholar] [CrossRef]
- Ristivojević, P.; Andrić, F.L.; Trifković, J.Đ.; Vovk, I.; Stanisavljević, L.Ž.; Tešić, Ž.L.; Milojković–Opsenica, D.M. Pattern recognition methods and multivariate image analysis in HPTLC fingerprinting of propolis extracts. J. Chemom. 2014, 28, 301–310. [Google Scholar] [CrossRef]
- Chasset, T.; Häbe, T.T.; Ristivojevic, P.; Morlock, G.E. Profiling and classification of French propolis by combined multivariate data analysis of planar chromatograms and scanning direct analysis in real time mass spectra. J. Chromatogr. A 2016, 1465, 197–204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cretu, G.C.; Morlock, G.E. Analysis of anthocyanins in powdered berry extracts by planarchromatography linked with bioassay and mass spectrometry. Food Chem. 2014, 146, 104–112. [Google Scholar] [CrossRef]
- Adhami, H.R.; Scherer, U.; Kaehlig, H.; Hettich, T.; Schlotterbeck, G.; Reich, E.; Krenn, L. Combination of bioautography with HPTLC–MS/NMR: A fast identification of acetylcholinesterase inhibitors from galbanum. Phytochem. Anal. 2013, 24, 395–400. [Google Scholar] [CrossRef] [PubMed]
- Shawky, E.; Sohafy, S.M.E. Untargeted and targeted chemical profiling for efficacy–directed discrimination of Hedera helix L. subspecies using HPTLC– image analysisand HPTLC/MS. Ind. Crops Prod. 2020, 145, 111980. [Google Scholar] [CrossRef]
- Fichou, D.; Ristivojević, P.; Morlock, G.E. Proof–of–principle of rTLC; an open–source software developed for image evaluation and multivariate analysis of planar chromatograms. Anal. Chem. 2016, 88, 12494–12501. [Google Scholar] [CrossRef] [Green Version]
- Salomé–Abarca, L.F.; Mandrone, M.; Sanna, C.; Poli, F.; van der Hondel, C.A.M.J.J.; Klinkhamer, P.G.L.; Choi, Y.H. Metabolic variation in Cistus monspeliensis L. ecotypes correlated to their plant–fungal interactions. Phytochemistry 2020, 176, e112402. [Google Scholar] [CrossRef]
- Pinu, F.R.; Villas–Boas, S.G.; Aggio, R. Analysis of intracellular metabolites from microorganisms: Quenching and extraction protocols. Metabolites 2017, 7, 53. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, H.S.; Delgado–Bertéli, M.B.; Cordeiro, F.A.; Lopes, A.D.; Silveira do Valle, J.; Linde, G.A.; Barros–Colauto, N. Semisolid culture medium improves mycelial recovery of Agaricussu brufescens cryo preserved in cereal grains. Braz. J. Microbiol. 2019, 50, 527–532. [Google Scholar] [CrossRef]
- Siddique, A.B.; Ebrahim, H.; Mohyeldin, M.; Qusa, M.; Batarseh, Y.; Fayyad, A.; Tajmim, A.; Nazzal, S.; Kaddoumi, A.; ElSayed, K. Novel liquid–liquid extraction and self–emulsion methods for simplified isolation of extra–virgin olive oil phenolics with emphasison (–)–oleocanthal and its oral anti–breast cancer activity. PLoS ONE 2019, 14, e0214798. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Charegaonkar, D. High-performance thin-layer chromatography: Excellent automation. In High-Performance Thin-Layer Chromatography (HPTLC), 1st ed.; Srivastava, M., Ed.; Springer: Berlin, Germany, 2011; pp. 55–65. [Google Scholar]
- Audoin, C.; Holderith, S.; Romari, K.; Thomas, O.P.; Genta-Jouve, G. Development of a work-flow for high-performance thin-layer chromatography data processing for untargeted metabolomics. J. Planar Chrom. 2014, 27, 328–332. [Google Scholar] [CrossRef]
- Saron-Eyob, O.; Parmar, S.; Wangab, Z.T.; Bligh, S.W.A. Metabolomics of four TCM herbal products: Application of HPTLC analysis. Anal. Methods 2012, 4, 2522. [Google Scholar]
- Lal, M.; Kadam, A.; Padole, A.S. HPTLC method development and validation of stigmasterol from different extracts of Tagetes erecta and Capsicum annuum. J. Drug Deliv. Ther. 2019, 9, 169–172. [Google Scholar] [CrossRef]
- Bhardwaj, A.; Pal, M.; Srivastava, M.; Tulsawani, R.; Sugadev, R.; Misra, K. HPTLC based chemometrics of medicinal mushrooms. J. Liq. Chromatogr. Relat. Technol. 2015, 38, 1392–1406. [Google Scholar] [CrossRef]
- Arruda-Frommenwiler, D.; Trefzer, D.; Schmid, M.; Cañigueral, S.; Reich, E. Comprehensive HPTLC fingerprinting: A novel economic approach to evaluating the quality of Ganoderma lucidum fruiting body. J. Liquid Chromatogr. Relat. Technol. 2020, 43, 414–423. [Google Scholar] [CrossRef]
- Orsini, F.; Vovk, I.; Glavnik, V.; Jug, U.; Corradini, D. HPTLC, HPTLC-MS/MS and HPTLC-DPPH methods for analyses of flavonoids and their antioxidant activity in Cyclanthera pedata leaves, fruits and dietary supplement. J. Liquid Chromatogr. Relat. Technol. 2019, 42, 290–301. [Google Scholar] [CrossRef]
- Osman, W.; Mohammed, M.S.; Khalid, H.S.; Muddathir, A.; Shantier, S.W.; Osman, B.; Abdoon, I. HPTLC Fingerprint Profile and Identification of Antidiabetic and Antioxidant Leads from Bauhinia rufescens L. Adv. Pharmacol. Pharm. Sci. 2020, 2020, 201463. [Google Scholar] [CrossRef] [Green Version]
- Ristivojević, P.; Dimkić, I.; Trifković, J.; Berić, T.; Vovk, I.; Milojković–Opsenica, D.; Stanković, S. Antimicrobial activity of Serbian propolis evaluated by means of MIC, HPTLC, bioautography and chemometrics. PLoS ONE 2016, 11, e0157097. [Google Scholar] [CrossRef] [PubMed]
- Darwish, R.S.; Shawky, E.; Hammoda, H.M.; Harraz, F.M. Peroxidase inhibitory and antioxidant constituents from Juniperus, L. species guided by HPTLC bioautography and molecular docking studies. Nat. Prod. Res. 2019, 9, 1–5. [Google Scholar] [CrossRef]
- Theiler, B.A.; Istvanits, S.; Zehl, M.; Marcourt, L.; Urban, E.; Espinoza–Caisa, L.O.; Glasl, S. HPTLC Bioautography Guided Isolation of α-Glucosidase Inhibiting Compounds from Justicia secunda Vahl (Acanthaceae). Phytochem. Anal. 2016, 28, 87–92. [Google Scholar] [CrossRef] [PubMed]
- Keller, N.P. Fungal secondary metabolism: Regulation, function and drug discovery. Nat. Rev. Microbiol. 2019, 17, 167–180. [Google Scholar] [CrossRef]
- Spiteller, P. Chemical defense strategies of higher fungi. Chem. Eur. J. 2008, 14, 9100–9110. [Google Scholar] [CrossRef]
- Waqas, M.; Khana, A.L.; Lee, I.J. Bioactive chemical constituents produced by endophytes and effects on rice plant growth. J. Plant Interact. 2014, 9, 478–487. [Google Scholar] [CrossRef]
- Bouyahya, A.; Dakka, N.; Et–Touys, A.; Abrini, J.; Bakri, Y. Medicinal plant products targeting quorum sensing for combating bacterial infections. Asian Pac. J. Trop. Med. 2017, 10, 729–743. [Google Scholar] [CrossRef]
- Maggini, V.; De Leo, M.; Mengoni, A.; Rosaria–Gallo, E.; Miceli, E.; Bandeira–Reidel, R.V.; Biffi, S.; Pistelli, L.; Fani, R.; Firenzuoli, F.; et al. Plant–endophytes interaction influences the secondary metabolism in Echinacea purpurea (L.) Moench: An in vitro model. Sci. Rep. 2017, 7, 16924. [Google Scholar] [CrossRef]
- Huang, L.H.; Yuan, M.Q.; Ao, X.J.; Ren, A.Y.; Zhang, H.B.; Yang, M.Z. Endophytic fungi specifically introduce novel metabolites into grape flesh cells in vitro. PLoS ONE 2018, 7, e0196996. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qin, D.; Wang, L.; Han, M.; Wang, J.; Song, H.; Yan, X.; Duan, X.; Dong, J. Effects of an endophytic fungus Umbelopsis dimorpha on the secondary metabolites of host–plant Kadsura angustifolia. Front. Microbiol. 2018, 9, 2845. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Combѐs, A.; Ndoye, I.; Bance, C.; Bruzaud, J.; Djediat, C.; Dupont, J.; Nay, B.; Prado, S. Chemical communication between the endophytic fungus Paraconiothyrium variabile and the phytopathogen Fusarium oxysporum. PLoS ONE 2012, 7, e47313. [Google Scholar] [CrossRef] [PubMed]
- Fernánndez–Conradi, P.; Jactel, H.E.; Robin, C.; Tack, A.J.M.; Castagneyrol, B. Fungi reduce preference and performance of insect herbivores on challenged plants. Ecology 2018, 99, 300–311. [Google Scholar] [CrossRef]
- Esser, K.; Meinhardt, F. Barrage Formation in Fungi. In Cellular Interactions—Encyclopedia of Plant Physiology, 17th ed.; Linskens, H.F., Heslop–Harrison, J., Eds.; Springer: Berlin/Heidelberg, Germany, 1984; pp. 350–354. [Google Scholar]
- Mishra, J.; Bhardwaj, A.; Pal, M.; Rajput, R.; Misr, K. High performance thin layer chromatography hyphenated with electrospray mass spectrometry for evaluation of nucleobases in two traditional Chinese medicinal mushrooms: A metabolomic approach. J. Liq. Chromatogr. Relat. Technol. 2018, 41, 910–918. [Google Scholar] [CrossRef]
- Jug, U.; Glavnik, V.; Kranjc, E.; Vovk, I. HPTLC–densitometric and HPTLC–MS methods for analysis of flavonoids. J. Liq. Chromatogr. Relat. Technol. 2018, 40, 329–341. [Google Scholar] [CrossRef]
- Yüce, I.; Mayr, M.; Morlock, G.E. Quantitative inkjet application on self-printed, binder-free HPTLC layers for submicromole-scaled analytical 1 H NMR spectroscopy. Anal. Chim. Acta 2019, 1087, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Van Kan, J.A.; Stassen, J.H.; Mosbach, A.; Van Der Lee, T.A.; Faino, L.; Farmer, A.D.; Papasotiriou, D.G.; Zhou, S.; Seidl, M.F.; Cottam, E.; et al. A gapless genome sequence of the fungus Botrytis cinerea. Mol. Plant Pathol. 2017, 18, 75–89. [Google Scholar] [CrossRef] [Green Version]
- Kurm, V.; van der Putten, W.H.; Hol, W.H.G. Cultivation-success of rare soil bacteria is not influenced by incubation time and growth medium. PLoS ONE 2019, 14, e0210073. [Google Scholar] [CrossRef]
- NCCLS. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically; Document M7-A6; NCCLS: Wayne, PA, USA, 2003. [Google Scholar]
- Abreu, A.C.; Serra, S.C.; Borges, A.; Saavedra, M.J.; Mcbain, A.J.; Salgado, A.J.; Simões, M. Combinatorial activity of flavonoids with antibiotics against drug-resistant Staphylococcus aureus. Microb. Drug Resist. 2015, 21, 600–609. [Google Scholar] [CrossRef] [PubMed]
- Arentshorst, M.; Ram, A.F.J.; Meyer, V. Using Non–homologous End–Joining–Deficient Strains for Functional Gene Analyses in Filamentous Fungi. In Plant Fungal Pathogens, 1st ed; Bolton, M., Thomma, B., Eds.; Springer: New York, NY, USA; Dordrecht, The Netherlands; Heidelberg, Germany; London, UK, 2012; pp. 133–150. [Google Scholar]







| Figure | Matched Identity | Primer | Query (%) | Similarity (%) |
|---|---|---|---|---|
| EU/PDA/ST/11GPC a | Colletotrichum godetiae | Forward | 88 | 99.82 |
| Colletotrichum acutatum | Forward | 88 | 99.82 | |
| Colletotrichum acutatum | Forward | 88 | 99.82 | |
| EU/PDA/ST/11GPC | Colletotrichum acutatum | Reverse | 94 | 99.01 |
| Colletotrichum acutatum | Reverse | 94 | 99.01 | |
| Colletotrichum acutatum | Reverse | 94 | 99.01 | |
| AS/PDA/5/3/2 b | Uncultured ascomycete | Forward | 92 | 99.85 |
| Penicillium crustosum | Forward | 92 | 99.85 | |
| Penicillium commune | Forward | 92 | 99.56 | |
| AS/PDA/5/3/2 | Uncultured ascomycete | Reverse | 93 | 98.99 |
| Penicillium crustosum | Reverse | 93 | 98.99 | |
| Penicillium crustosum | Reverse | 93 | 98.99 | |
| AS/PDA/4/3/2 c | Uncultured Penicillium | Forward | 71 | 99.49 |
| Talaromyces purpurogenus | Forward | 69 | 99.47 | |
| Fungal endophyte SPSX01 | Forward | 69 | 99.47 | |
| AS/PDA/4/3/2 | Uncultured Penicillium | Reverse | 77 | 99.83 |
| Talaromyces purpurogenus | Reverse | 75 | 99.83 | |
| Fungal endophyte SPSX01 | Reverse | 75 | 99.83 | |
| AS/PDA/11(2) d | Uncultured Penicillium | Forward | 71 | 98.29 |
| Talaromyces purpurogenus | Forward | 70 | 98.25 | |
| Fungal endophyte SPSX01 | Forward | 70 | 98.25 | |
| AS/PDA/11(2) | Uncultured Penicillium | Reverse | 88 | 98.84 |
| Talaromyces albobiverticillius | Reverse | 81 | 98.66 | |
| Talaromyces purpurogenus | Reverse | 80 | 92.82 |
| Primer’s Name | Sense | Sequence 5′-3′ | Gene |
|---|---|---|---|
| V9g | Forward | TTACGTCCCTGCCCTTTGTA | ITS1 and ITS2 |
| ITS4 | Reverse | TCCTCCGCTTATTGATATGC | regions + 5.8S |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Salomé-Abarca, L.F.; van den Hondel, C.A.M.J.J.; Erol, Ö.; Klinkhamer, P.G.L.; Kim, H.K.; Choi, Y.H. HPTLC-Based Chemical Profiling: An Approach to Monitor Plant Metabolic Expansion Caused by Fungal Endophytes. Metabolites 2021, 11, 174. https://doi.org/10.3390/metabo11030174
Salomé-Abarca LF, van den Hondel CAMJJ, Erol Ö, Klinkhamer PGL, Kim HK, Choi YH. HPTLC-Based Chemical Profiling: An Approach to Monitor Plant Metabolic Expansion Caused by Fungal Endophytes. Metabolites. 2021; 11(3):174. https://doi.org/10.3390/metabo11030174
Chicago/Turabian StyleSalomé-Abarca, Luis F., Cees A. M. J. J. van den Hondel, Özlem Erol, Peter G. L. Klinkhamer, Hye Kyong Kim, and Young Hae Choi. 2021. "HPTLC-Based Chemical Profiling: An Approach to Monitor Plant Metabolic Expansion Caused by Fungal Endophytes" Metabolites 11, no. 3: 174. https://doi.org/10.3390/metabo11030174
APA StyleSalomé-Abarca, L. F., van den Hondel, C. A. M. J. J., Erol, Ö., Klinkhamer, P. G. L., Kim, H. K., & Choi, Y. H. (2021). HPTLC-Based Chemical Profiling: An Approach to Monitor Plant Metabolic Expansion Caused by Fungal Endophytes. Metabolites, 11(3), 174. https://doi.org/10.3390/metabo11030174