Performance Verification of CYP2C19 Enzyme Abundance Polymorphism Settings within the Simcyp Simulator v21
Abstract
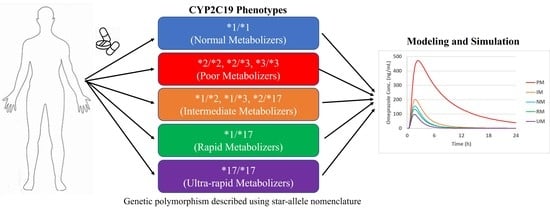
:1. Introduction
2. Materials and Methods
2.1. Literature Search
2.2. Modeling Strategy
- Recovery of area under the concentration-time curve (AUC) and maximum concentration (Cmax) from compiled datasets
- Recovery of victim DDI ratios with CYP2C19 and/or CYP3A4 inhibition
- Recovery of the relative change in exposure between phenotypes compared to NM, where available, from select studies
2.3. Omeprazole and Lansoprazole PK Prediction in CYP2C19 Phenotypes
2.4. Victim DDI Prediction in CYP2C19 Phenotypes
2.5. Prediction of CYP2C19 Phenotype Ratios
3. Results
3.1. Literature Search
3.2. Omeprazole and Lansoprazole PK Prediction in CYP2C19 Phenotypes
3.3. Victim DDI Prediction in CYP2C19 Phenotypes
3.4. Prediction of CYP2C19 Phenotype Ratios
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bahar, M.A.; Setiawan, D.; Hak, E.; Wilffert, B. Pharmacogenetics of drug-drug interaction and drug-drug-gene interaction: A systematic review on CYP2C9, CYP2C19 and CYP2D6. Pharmacogenomics 2017, 18, 701–739. [Google Scholar] [CrossRef] [PubMed]
- US Food and Drug Administration. Guidance for Industry: Clinical Drug Interaction Studies—Cytochrome P450 Enzyme- and Transporter-Mediated Drug Interactions. Available online: https://www.fda.gov/media/134581/download (accessed on 14 June 2022).
- European Medicines Agency. Guideline on the Investigation of Drug Interactions. Available online: https://www.ema.europa.eu/en/documents/scientific-guideline/guideline-investigation-drug-interactions-revision-1_en.pdf (accessed on 14 June 2022).
- Zhang, X.; Yang, Y.; Grimstein, M.; Fan, J.; Grillo, J.A.; Huang, S.M.; Zhu, H.; Wang, Y. Application of PBPK Modeling and Simulation for Regulatory Decision Making and Its Impact on US Prescribing Information: An Update on the 2018-2019 Submissions to the US FDA’s Office of Clinical Pharmacology. J. Clin. Pharmacol. 2020, 60 (Suppl. 1), S160–S178. [Google Scholar] [CrossRef]
- Rostami-Hodjegan, A. Reverse translation in PBPK and QSP: Going backwards in order to go forward with confidence. Clin. Pharmacol. Ther. 2018, 103, 224–232. [Google Scholar] [CrossRef] [Green Version]
- Zhao, M.; Ma, J.; Li, M.; Zhang, Y.; Jiang, B.; Zhao, X.; Huai, C.; Shen, L.; Zhang, N.; He, L.; et al. Cytochrome P450 enzymes and drug metabolism in humans. Int. J. Mol. Sci. 2021, 22, 12808. [Google Scholar] [CrossRef] [PubMed]
- Fanni, D.; Pinna, F.; Gerosa, C.; Paribello, P.; Carpiniello, B.; Faa, G.; Manchia, M. Anatomical distribution and expression of CYP in humans: Neuropharmacological implications. Drug Dev Res. 2021, 85, 628–667. [Google Scholar] [CrossRef] [PubMed]
- Manikandan, P.; Nagini, S. Cytochrome P450 structure, function and clinical significance: A review. Curr Drug Targets 2018, 19, 38–54. [Google Scholar] [CrossRef] [PubMed]
- Zanger, U.M.; Schwab, M. Cytochrome P450 enzymes in drug metabolism: Regulation of gene expression, enzyme activities, and impart of genetic variation. Pharmacol. Ther. 2013, 138(1), 103–141. [Google Scholar] [CrossRef]
- Klein, M.D.; Williams, A.K.; Lee, C.R.; Stouffer, G.A. Clinical Utility of CYP2C19 Genotyping to Guide Antiplatelet Therapy in Patients With an Acute Coronary Syndrome or Undergoing Percutaneous Coronary Intervention. Arterioscler. Thromb. Vasc. Bio. 2019, 39(4), 647–652. [Google Scholar] [CrossRef] [Green Version]
- Ozawa, S.; Soyama, A.; Saeki, M.; Fukushima-Uesaka, H.; Itoda, M.; Koyano, S.; Sai, K.; Ohno, Y.; Saito, Y.; Sawada, J. Ethnic differences in genetic polymorphisms of CYP2D6, CYP2C19, CYP3As and MDR1/ABCB1. Drug Metab. Pharmacokinet. 2004, 19, 83–95. [Google Scholar] [CrossRef]
- Kita, T.; Sakaeda, T.; Baba, T.; Aoyama, N.; Kakumoto, M.; Kurimoto, Y.; Kawahara, Y.; Okamura, N.; Kirita, S.; Kasuga, M.; et al. Different contribution of CYP2C19 in the in vitro metabolism of three proton pump inhibitors. Biol. Pharm. Bull. 2003, 26, 386–390. [Google Scholar] [CrossRef]
- US Food and Drug Administration. Drug Development and Drug Interactions|Table of Substrates, Inhibitors and Inducers. Available online: https://www.fda.gov/drugs/drug-interactions-labeling/drug-development-and-drug-interactions-table-substrates-inhibitors-and-inducers#table2-1 (accessed on 14 June 2022).
- University of Washington Drug Interaction Solutions. Available online: https://www.druginteractionsolutions.org/ (accessed on 16 November 2021).
- Anil. Digitize2.m. MATLAB Central File Exchange. 2022. Available online: https://www.mathworks.com/matlabcentral/fileexchange/928-digitize2-m (accessed on 29 June 2021).
- Zhou, L.; Sharma, P.; Yeo, K.R.; Higashimori, M.; Xu, H.; Al-Huniti, N.; Zhou, D. Assessing pharmacokinetic differences in Caucasian and East Asian (Japanese, Chinese and Korean) populations driven by CYP2C19 polymorphism using physiologically-based pharmacokinetic modelling. Eur. J. Pharm. Sci. 2019, 139, 105061. [Google Scholar] [CrossRef] [PubMed]
- Zvyaga, T.; Chang, S.Y.; Chen, C.; Yang, Z.; Vuppugalla, R.; Hurley, J.; Thorndike, D.; Wagner, A.; Chimalakonda, A.; Rodrigues, A.D. Evaluation of six proton pump inhibitors as inhibitors of various human cytochromes P450: Focus on cytochrome P450 2C19. Drug Metab. Dispos. 2012, 40, 1698–1711. [Google Scholar] [CrossRef] [Green Version]
- Baldwin, R.M.; Ohlsson, S.; Pedersen, R.S.; Mwinyi, J.; Ingelman-Sundberg, M.; Eliasson, E.; Bertilsson, L. Increased omeprazole metabolism in carriers of the CYP2C19*17 allele; a pharmacokinetic study in healthy volunteers. Br. J. Clin. Pharmacol. 2008, 65, 767–774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, M.; Tybring, G.; Dahl, M.L.; Gotharson, E.; Sagar, M.; Seensalu, R.; Bertilsson, L. Interphenotype differences in disposition and effect on gastrin levels of omeprazole--suitability of omeprazole as a probe for CYP2C19. Br. J. Clin. Pharmacol. 1995, 39, 511–518. [Google Scholar] [CrossRef] [Green Version]
- Cho, J.Y.; Yu, K.S.; Jang, I.J.; Yang, B.H.; Shin, S.G.; Yim, D.S. Omeprazole hydroxylation is inhibited by a single dose of moclobemide in homozygotic EM genotype for CYP2C19. Br. J. Clin. Pharmacol. 2002, 53, 393–397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Furuta, T.; Ohashi, K.; Kobayashi, K.; Iida, I.; Yoshida, H.; Shirai, N.; Takashima, M.; Kosuge, K.; Hanai, H.; Chiba, K.; et al. Effects of clarithromycin on the metabolism of omeprazole in relation to CYP2C19 genotype status in humans. Clin. Pharmacol. Ther. 1999, 66, 265–274. [Google Scholar] [CrossRef]
- Furuta, T.; Ohashi, K.; Kosuge, K.; Zhao, X.J.; Takashima, M.; Kimura, M.; Nishimoto, M.; Hanai, H.; Kaneko, E.; Ishizaki, T. CYP2C19 genotype status and effect of omeprazole on intragastric pH in humans. Clin. Pharmacol. Ther. 1999, 65, 552–561. [Google Scholar] [CrossRef]
- Furuta, T.; Shirai, N.; Xiao, F.; Ohashi, K.; Ishizaki, T. Effect of high-dose lansoprazole on intragastic pH in subjects who are homozygous extensive metabolizers of cytochrome P4502C19. Clin. Pharmacol. Ther. 2001, 70, 484–492. [Google Scholar] [CrossRef]
- He, N.; Huang, S.L.; Zhu, R.H.; Tan, Z.R.; Liu, J.; Zhu, B.; Zhou, H.H. Inhibitory effect of troleandomycin on the metabolism of omeprazole is CYP2C19 genotype-dependent. Xenobiotica 2003, 33, 211–221. [Google Scholar] [CrossRef]
- Hu, X.P.; Xu, J.M.; Hu, Y.M.; Mei, Q.; Xu, X.H. Effects of CYP2C19 genetic polymorphism on the pharmacokinetics and pharmacodynamics of omeprazole in Chinese people. J. Clin. Pharm. Ther. 2007, 32, 517–524. [Google Scholar] [CrossRef]
- Hunfeld, N.G.; Mathot, R.A.; Touw, D.J.; van Schaik, R.H.; Mulder, P.G.; Franck, P.F.; Kuipers, E.J.; Geus, W.P. Effect of CYP2C19*2 and *17 mutations on pharmacodynamics and kinetics of proton pump inhibitors in Caucasians. Br. J. Clin. Pharmacol. 2008, 65, 752–760. [Google Scholar] [CrossRef] [PubMed]
- Ieiri, I.; Kubota, T.; Urae, A.; Kimura, M.; Wada, Y.; Mamiya, K.; Yoshioka, S.; Irie, S.; Amamoto, T.; Nakamura, K.; et al. Pharmacokinetics of omeprazole (a substrate of CYP2C19) and comparison with two mutant alleles, C gamma P2C19m1 in exon 5 and C gamma P2C19m2 in exon 4, in Japanese subjects. Clin. Pharmacol. Ther. 1996, 59, 647–653. [Google Scholar] [CrossRef]
- Ieiri, I.; Kishimoto, Y.; Okochi, H.; Momiyama, K.; Morita, T.; Kitano, M.; Morisawa, T.; Fukushima, Y.; Nakagawa, K.; Hasegawa, J.; et al. Comparison of the kinetic disposition of and serum gastrin change by lansoprazole versus rabeprazole during an 8-day dosing scheme in relation to CYP2C19 polymorphism. Eur. J. Clin. Pharmacol. 2001, 57, 485–492. [Google Scholar] [CrossRef]
- Ieiri, I.; Kimura, M.; Irie, S.; Urae, A.; Otsubo, K.; Ishizaki, T. Interaction magnitude, pharmacokinetics and pharmacodynamics of ticlopidine in relation to CYP2C19 genotypic status. Pharmacogenet. Genomics 2005, 15, 851–859. [Google Scholar] [CrossRef] [PubMed]
- Ishizawa, Y.; Yasui-Furukori, N.; Takahata, T.; Sasaki, M.; Tateishi, T. The effect of aging on the relationship between the cytochrome P450 2C19 genotype and omeprazole pharmacokinetics. Clin. Pharmacokinet. 2005, 44, 1179–1189. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.K.; Kang, T.S.; Eom, S.O.; Kim, J.I.; Lee, H.J.; Roh, J. CYP2C19 haplotypes in Koreans as a marker of enzyme activity evaluated with omeprazole. J. Clin. Pharm. Ther. 2009, 34, 437–446. [Google Scholar] [CrossRef] [PubMed]
- Li, C.Y.; Zhang, J.; Chu, J.H.; Xu, M.J.; Ju, W.Z.; Liu, F.; Jian-Dong, Z. A correlative study of polymorphisms of CYP2C19 and MDR1 C3435T with the pharmacokinetic profiles of lansoprazole and its main metabolites following single oral administration in healthy adult Chinese subjects. Eur. J. Drug Metab. Pharmacokinet. 2014, 39, 121–128. [Google Scholar] [CrossRef]
- Michaud, V.; Kreutz, Y.; Skaar, T.; Ogburn, E.; Thong, N.; Flockhart, D.A.; Desta, Z. Efavirenz-mediated induction of omeprazole metabolism is CYP2C19 genotype dependent. Pharmacogenomics J. 2014, 14, 151–159. [Google Scholar] [CrossRef] [Green Version]
- Miura, M.; Tada, H.; Yasui-Furukori, N.; Uno, T.; Sugawara, K.; Tateishi, T.; Suzuki, T. Enantioselective disposition of lansoprazole in relation to CYP2C19 genotypes in the presence of fluvoxamine. Br. J. Clin. Pharmacol. 2005, 60, 61–68. [Google Scholar] [CrossRef]
- Nazir, S.; Iqbal, Z.; Ahmad, L.; Shah, Y.; Nasir, F. Pharmacokinetics of omeprazole and its metabolites in three phases of menstrual cycle. Eur. J. Drug Metab. Pharmacokinet. 2015, 40, 13–22. [Google Scholar] [CrossRef]
- Nazir, S.; Iqbal, Z.; Ahmad, L.; Ahmad, S. Variation in Pharmacokinetics of Omeprazole and Its Metabolites by Gender and CYP2C19 Genotype in Pakistani Male and Female Subjects. Pak. J. Pharm. Sci. 2016, 29, 887–894. Available online: https://www.ncbi.nlm.nih.gov/pubmed/27166533 (accessed on 1 September 2022). [PubMed]
- Park, S.; Hyun, Y.J.; Kim, Y.R.; Lee, J.H.; Ryu, S.; Kim, J.M.; Oh, W.Y.; Na, H.S.; Lee, J.G.; Seo, D.W.; et al. Effects of CYP2C19 Genetic Polymorphisms on PK/PD Responses of Omeprazole in Korean Healthy Volunteers. J. Korean Med. Sci. 2017, 32, 729–736. [Google Scholar] [CrossRef] [PubMed]
- Payan, M.; Rouini, M.R.; Tajik, N.; Ghahremani, M.H.; Tahvilian, R. Hydroxylation index of omeprazole in relation to CYP2C19 polymorphism and sex in a healthy Iranian population. Daru 2014, 22, 81. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qiao, H.L.; Hu, Y.R.; Tian, X.; Jia, L.J.; Gao, N.; Zhang, L.R.; Guo, Y.Z. Pharmacokinetics of three proton pump inhibitors in Chinese subjects in relation to the CYP2C19 genotype. Eur. J. Clin. Pharmacol. 2006, 62, 107–112. [Google Scholar] [CrossRef]
- Roman, M.; Ochoa, D.; Sanchez-Rojas, S.D.; Talegon, M.; Prieto-Perez, R.; Rivas, A.; Abad-Santos, F.; Cabaleiro, T. Evaluation of the relationship between polymorphisms in CYP2C19 and the pharmacokinetics of omeprazole, pantoprazole and rabeprazole. Pharmacogenomics 2014, 15, 1893–1901. [Google Scholar] [CrossRef]
- Saito, M.; Yasui-Furukori, N.; Uno, T.; Takahata, T.; Sugawara, K.; Munakata, A.; Tateishi, T. Effects of clarithromycin on lansoprazole pharmacokinetics between CYP2C19 genotypes. Br. J. Clin. Pharmacol. 2005, 59, 302–309. [Google Scholar] [CrossRef] [Green Version]
- Sakai, T.; Aoyama, N.; Kita, T.; Sakaeda, T.; Nishiguchi, K.; Nishitora, Y.; Hohda, T.; Sirasaka, D.; Tamura, T.; Tanigawara, Y.; et al. CYP2C19 genotype and pharmacokinetics of three proton pump inhibitors in healthy subjects. Pharm. Res. 2001, 18, 721–727. [Google Scholar] [CrossRef]
- Shimizu, M.; Uno, T.; Niioka, T.; Yaui-Furukori, N.; Takahata, T.; Sugawara, K.; Tateishi, T. Sensitive determination of omeprazole and its two main metabolites in human plasma by column-switching high-performance liquid chromatography: Application to pharmacokinetic study in relation to CYP2C19 genotypes. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2006, 832, 241–248. [Google Scholar] [CrossRef]
- Shiohira, H.; Yasui-Furukori, N.; Yamada, S.; Tateishi, T.; Akamine, Y.; Uno, T. Hydroxylation of R(+)- and S(-)-omeprazole after racemic dosing are different among the CYP2C19 genotypes. Pharm. Res. 2012, 29, 2310–2316. [Google Scholar] [CrossRef]
- Shirai, N.; Furuta, T.; Moriyama, Y.; Okochi, H.; Kobayashi, K.; Takashima, M.; Xiao, F.; Kosuge, K.; Nakagawa, K.; Hanai, H.; et al. Effects of CYP2C19 genotypic differences in the metabolism of omeprazole and rabeprazole on intragastric pH. Aliment. Pharmacol. Ther. 2001, 15, 1929–1937. [Google Scholar] [CrossRef]
- Tu, J.H.; Hu, D.L.; Dai, L.L.; Sun, Y.; Fan, L.; Zhang, M.; Tan, Z.R.; Chen, Y.; Li, Z.; Zhou, H.H. Effect of glycyrrhizin on CYP2C19 and CYP3A4 activity in healthy volunteers with different CYP2C19 genotypes. Xenobiotica 2010, 40, 393–399. [Google Scholar] [CrossRef] [PubMed]
- Uno, T.; Yasui-Furukori, N.; Takahata, T.; Sugawara, K.; Tateishi, T. Lack of significant effect of grapefruit juice on the pharmacokinetics of lansoprazole and its metabolites in subjects with different CYP2C19 genotypes. J. Clin. Pharmacol. 2005, 45, 690–694. [Google Scholar] [CrossRef]
- Uno, T.; Niioka, T.; Hayakari, M.; Yasui-Furukori, N.; Sugawara, K.; Tateishi, T. Absolute bioavailability and metabolism of omeprazole in relation to CYP2C19 genotypes following single intravenous and oral administrations. Eur. J. Clin. Pharmacol. 2007, 63, 143–149. [Google Scholar] [CrossRef]
- Xu, H.R.; Chen, W.L.; Li, X.N.; Chu, N.N. The effect of CYP2C19 activity on pharmacokinetics of lansoprazole and its active metabolites in healthy subjects. Pharm. Biol. 2010, 48, 947–952. [Google Scholar] [CrossRef] [PubMed]
- Yamada, S.; Shiohira, H.; Yasui-Furukori, N.; Tateishi, T.; Akamine, Y.; Uno, T. The (R)-omeprazole hydroxylation index reflects CYP2C19 activity in healthy Japanese volunteers. Eur. J. Clin. Pharmacol. 2013, 69, 1423–1428. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.J.; Fan, L.; Liu, Z.Q.; Mao, Y.M.; Guo, D.; Liu, L.H.; Tan, Z.R.; Peng, L.; Han, C.T.; Hu, D.L.; et al. Effects of allicin on CYP2C19 and CYP3A4 activity in healthy volunteers with different CYP2C19 genotypes. Eur. J. Clin. Pharmacol. 2009, 65, 601–608. [Google Scholar] [CrossRef]
- Yasui-Furukori, N.; Saito, M.; Uno, T.; Takahata, T.; Sugawara, K.; Tateishi, T. Effects of fluvoxamine on lansoprazole pharmacokinetics in relation to CYP2C19 genotypes. J. Clin. Pharmacol. 2004, 44, 1223–1229. [Google Scholar] [CrossRef]
- Yin, O.Q.; Tomlinson, B.; Chow, A.H.; Waye, M.M.; Chow, M.S. Omeprazole as a CYP2C19 marker in Chinese subjects: Assessment of its gene-dose effect and intrasubject variability. J. Clin. Pharmacol. 2004, 44, 582–589. [Google Scholar] [CrossRef]
- Zhang, D.; Wang, X.; Yang, M.; Wang, G.; Liu, H. Effects of CYP2C19 polymorphism on the pharmacokinetics of lansoprazole and its main metabolites in healthy Chinese subjects. Xenobiotica 2011, 41, 511–517. [Google Scholar] [CrossRef]
- Landes, B.D.; Petite, J.P.; Flouvat, B. Clinical pharmacokinetics of lansoprazole. Clin. Pharmacokinet. 1995, 28, 458–470. [Google Scholar] [CrossRef]
- Andersson, T.; Cederberg, C.; Regardh, C.G.; Skanberg, I. Pharmacokinetics of various single intravenous and oral doses of omeprazole. Eur. J. Clin. Pharmacol. 1990, 39, 195–197. [Google Scholar] [CrossRef]
- Kim, Y.; Hatley, O.; Rhee, S.J.; Yi, S.; Lee, H.A.; Yoon, S.; Chung, J.Y.; Yu, K.S.; Lee, H. Development of a Korean-specific virtual population for physiologically based pharmacokinetic modelling and simulation. Biopharm. Drug Dispos. 2019, 40, 135–150. [Google Scholar] [CrossRef] [PubMed]
- Guest, E.J.; Aarons, L.; Houston, J.B.; Rostami-Hodjegan, A.; Galetin, A. Critique of the two-fold measure of prediction success for ratios: Application for the assessment of drug-drug interactions. Drug Metab. Dispos. 2011, 39, 170–173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wedlund, P.J.; Aslanian, W.S.; Jacqz, E.; McAllister, C.B.; Branch, R.A.; Wilkinson, G.R. Phenotypic Differences in Mephenytoin Pharmacokinetics in Normal Subjects. J. Pharmacol. Exp. Ther. 1985, 234, 662–669. Available online: https://www.ncbi.nlm.nih.gov/pubmed/4032286 (accessed on 1 September 2022). [PubMed]
- Tiwari, K.; Kananathan, S.; Roberts, M.G.; Meyer, J.P.; Sharif Shohan, M.U.; Xavier, A.; Maire, M.; Zyoud, A.; Men, J.; Ng, S.; et al. Reproducibility in systems biology modelling. Mol. Syst. Biol. 2021, 17, e9982. [Google Scholar] [CrossRef] [PubMed]



| Hepatic Abundance (pmol/mg protein) (CV%) | |||||
| Simcyp | PM | IM | NM | RM | UM |
| V21 | 0 | 2.85 (52) | 4.4 (52) | 7.01 (89) | 10.23 (79) |
| V20 | 0 | NA | 4.4 (71) | NA | 8.7 (71) |
| Intestinal Abundance (nmol/small intestine) (CV%) | |||||
| Simcyp | PM | IM | NM | RM | UM |
| V21 | 0 | 1.29 (52) | 2 (52) | 3.18 (89) | 4.65 (79) |
| V20 | 0 | NA | 2 (77) | NA | 4 (77) |
| No. of Simulated within 2-Fold of Observed/Total No. Data Points | |||||
|---|---|---|---|---|---|
| Omeprazole 1 | PM | IM | NM | RM | UM |
| AUC | 25/26 | 22/26 | 24/27 | 2/2 | 3/3 |
| Cmax | 12/17 | 8/16 | 10/17 | 0/1 | 1/2 |
| Lansoprazole 1 | |||||
| AUC | 7/10 | 10/11 | 11/11 | 1/1 | NA |
| Cmax | 9/10 | 9/10 | 9/10 | NA | NA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sychterz, C.; Gardner, I.; Chiang, M.; Rachumallu, R.; Neuhoff, S.; Perera, V.; Merali, S.; Schmidt, B.J.; Gaohua, L. Performance Verification of CYP2C19 Enzyme Abundance Polymorphism Settings within the Simcyp Simulator v21. Metabolites 2022, 12, 1001. https://doi.org/10.3390/metabo12101001
Sychterz C, Gardner I, Chiang M, Rachumallu R, Neuhoff S, Perera V, Merali S, Schmidt BJ, Gaohua L. Performance Verification of CYP2C19 Enzyme Abundance Polymorphism Settings within the Simcyp Simulator v21. Metabolites. 2022; 12(10):1001. https://doi.org/10.3390/metabo12101001
Chicago/Turabian StyleSychterz, Caroline, Iain Gardner, Manting Chiang, Ramakrishna Rachumallu, Sibylle Neuhoff, Vidya Perera, Samira Merali, Brian J. Schmidt, and Lu Gaohua. 2022. "Performance Verification of CYP2C19 Enzyme Abundance Polymorphism Settings within the Simcyp Simulator v21" Metabolites 12, no. 10: 1001. https://doi.org/10.3390/metabo12101001
APA StyleSychterz, C., Gardner, I., Chiang, M., Rachumallu, R., Neuhoff, S., Perera, V., Merali, S., Schmidt, B. J., & Gaohua, L. (2022). Performance Verification of CYP2C19 Enzyme Abundance Polymorphism Settings within the Simcyp Simulator v21. Metabolites, 12(10), 1001. https://doi.org/10.3390/metabo12101001