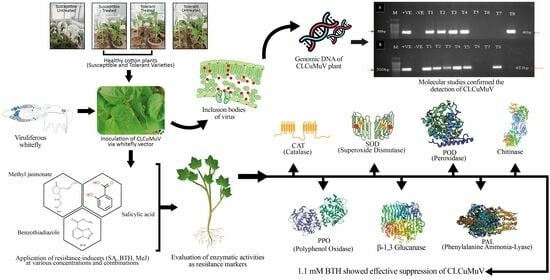
Elicitor-Driven Defense Mechanisms: Shielding Cotton Plants against the Onslaught of Cotton Leaf Curl Multan Virus (CLCuMuV) Disease
Abstract
:1. Introduction
2. Materials and Methods
2.1. Planting Material
2.2. Application of Elicitors (Uninoculated)
2.3. Rearing of A-Viruliferous Whiteflies
2.4. Application of Elicitors Followed by Virus Inoculation and Evaluation of Disease Severity
2.5. DNA Extraction and Confirmation of CLCuMuV Amplification through Polymerase Chain Reaction (PCR)
2.6. Determination of Enzymatic Activities in Order to Observe Biological Changes in Plants
2.7. Enzyme Extract for SOD, POD, and CAT
2.8. Assay for SOD (EC 1.15.1.1)
2.9. Assay for POD (EC 1.11.1.7)
2.10. Assay for CAT (EC 1.11.1.6)
2.11. Enzyme Extract and Assay for PAL (EC.4.3.1.24)
2.12. Enzyme Extract and Assay for PPO (EC. 1.14.18.1)
2.13. Enzyme Extraction for β–1,3-Glucanase and Chitinase and Colloidal Chitin for Chitinase Activity
2.14. Microplate Assay for β–1,3-Glucanase and Chitinase
2.15. Estimation of Total Phenolic Contents (EC 1.14.13.7)
2.16. Bradford Protein Assay
2.17. Correlation, Principal Component and Statistical Analysis
3. Results
3.1. Impact of Elicitors Treatments on Disease Severity of Cotton Leaf Curl Virus
3.2. Confirmation of CLCuMuV Infection through PCR
3.3. Impact of Elicitors Treatments on Defense-Related Enzymatic Activity of Cotton Plants
3.3.1. Superoxide Dismutase (SOD) Enzymatic Activity
3.3.2. Peroxidase (POD) Enzymatic Activity
3.3.3. Catalase (CAT) Enzymatic Activity
3.3.4. Polyphenoloxidase (PPO) Enzymatic Activity
3.3.5. Phenylalanine Amonialyase (PAL) Enzymatic Activity
3.3.6. Phenolic Activity
3.3.7. Beta 1,3 Glucanase Activity
3.3.8. Chitinase Activity
3.3.9. Effect of Different Elicitor Treatments on Overall Enzymatic Activities
3.4. Correlation Analysis among the Enzymatic Activities and Disease Severity
3.5. Principal Component Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Akhtar, K.P.; Khan, A.I.; Hussain, M.; Khan, M. Comparison of resistance level to cotton leaf curl virus (CLCuV) among newly developed cotton mutants and commercial cultivars. Plant Pathol. J. 2002, 18, 179–186. [Google Scholar] [CrossRef]
- Hameed, S.; Khalid, S.; Ehsan-ul-Haq, S.; Hashrni, A. Cotton leaf curl disease in Pakistan caused by a whitefly-transmitted geminivirus. Plant Dis. 1994, 78, 529. [Google Scholar] [CrossRef]
- Mansoor, S.; Bedford, I.; Pinner, M.; Stanley, J.; Markham, P. A whitefly-transmitted geminivirus associated with cotton leaf curl disease in Pakistan. Pak. J. Bot. 1993, 25, 105–107. [Google Scholar]
- Yassin, A.; El Nur, E. Transmission of Cotton leaf curl virus by single insects of Bemisia tabaci. Plant Dis. Report. 1970, 54, 528–531. [Google Scholar]
- Hemmati, F.; Behjatnia, S.A.-A.; Moghadam, A.; Afsharifar, A. Induction of systemic resistance against cucumber mosaic virus (CMV) and tomato yellow leaf curl virus (TYLCV) in tomato. Int. J. Pest Manag. 2023, 1–14. [Google Scholar] [CrossRef]
- Aqueel, R.; Badar, A.; Roy, N.; Mushtaq, Q.; Ali, A.F.; Bashir, A.; Ijaz, U.Z.; Malik, K.A. Cotton Microbiome Profiling and Cotton Leaf Curl Disease (CLCuD) Suppression through Microbial Consortia associated with Gossypium arboreum. bioRxiv 2023. [Google Scholar] [CrossRef]
- Mahmood, T.; Arshad, M.; Gill, M.I.; Mahmood, H.T.; Tahir, M.; Hussain, S. Burewala Strain of Cotton Leaf Curl Virus: A Threat to CLCuV Cotton Resistant Varieties. Asian J. Plant Sci. 2003, 2, 968–970. [Google Scholar] [CrossRef]
- Mansoor, S.; Briddon, R.; Bull, S.; Bedford, I.; Bashir, A.; Hussain, M.; Saeed, M.; Zafar, Y.; Malik, K.; Fauquet, C. Cotton leaf curl disease is associated with multiple monopartite begomoviruses supported by single DNA β. Arch. Virol. 2003, 148, 1969–1986. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, P.; Mehta, N.; Chauhan, R.S.; Kumar, A.; Singh, H.; Lal, M.K.; Tiwari, R.K.; Kumar, R. Utilization of primary and secondary biochemical compounds in cotton as diagnostic markers for measuring resistance to cotton leaf curl virus. Front. Plant Sci. 2023, 14, 1185337. [Google Scholar] [CrossRef]
- Atiq, M.; Talib, M.Z.; Rajput, N.A.; Sahi, S.T.; Usman, M.; Jabar, A.; Arif, M.J.; Gogi, M.D.; Khan, M.A.; Akram, A. NEW Fangled Tactics Towards Cotton Leaf Curl Virus Disease A Review. J. Nat. Fibers 2023, 20, 2217364. [Google Scholar] [CrossRef]
- Briddon, R.W.; E Bull, S.; Amin, I.; Idris, A.M.; Mansoor, S.; Bedford, I.D.; Dhawan, P.; Rishi, N.; Siwatch, S.S.; Abdel-Salam, A.M.; et al. Diversity of DNA β, a satellite molecule associated with some monopartite begomoviruses. Virology 2003, 312, 106–121. [Google Scholar] [CrossRef]
- Mao, M.-J.; He, Z.-F.; Yu, H.; Li, H.-P. Molecular characterization of cotton leaf Curl Multan virus and its satellite DNA that infects Hibiscus rosa-sinensis. Chin. J. Virol. 2008, 24, 64–68. [Google Scholar]
- Shine, M.B.; Xiao, X.; Kachroo, P.; Kachroo, A. Signaling mechanisms underlying systemic acquired resistance to microbial pathogens. Plant Sci. 2019, 279, 81–86. [Google Scholar] [CrossRef] [PubMed]
- Heil, M.; Bostock, R.M. Induced Systemic Resistance (ISR) Against Pathogens in the Context of Induced Plant Defences. Ann. Bot. 2002, 89, 503–512. [Google Scholar] [CrossRef]
- Pasquer, F.; Isidore, E.; Zarn, J.; Keller, B. Specific patterns of changes in wheat gene expression after treatment with three antifungal compounds. Plant Mol. Biol. 2005, 57, 693–707. [Google Scholar] [CrossRef]
- Appu, M.; Ramalingam, P.; Sathiyanarayanan, A.; Huang, J. An overview of plant defense-related enzymes responses to biotic stresses. Plant Gene 2021, 27, 100302. [Google Scholar] [CrossRef]
- Confortin, T.C.; Spannemberg, S.S.; Todero, I.; Luft, L.; Brun, T.; Alves, E.A.; Kuhn, R.C.; Mazutti, M.A. Microbial enzymes as control agents of diseases and pests in organic agriculture. In New and Future Developments in Microbial Biotechnology and Bioengineering; Elsevier: Amsterdam, The Netherlands, 2018; pp. 321–332. [Google Scholar]
- Garmendia, I.; Aguirreolea, J.; Goicoechea, N. Defence-related enzymes in pepper roots during interactions with arbuscular mycorrhizal fungi and/or Verticillium dahliae. BioControl 2006, 51, 293. [Google Scholar] [CrossRef]
- Zhang, J.; Stewart, J.M.D. Economical and Rapid Method for Extracting Cotton Genomic DNA. J. Cotton Sci. 2000, 4, 193–201. [Google Scholar]
- Idris, A.; Al-Saleh, M.; Amer, M.; Abdalla, O.; Brown, J. Introduction of Cotton leaf curl Gezira virus into the United Arab Emirates. Plant Dis. 2014, 98, 1593. [Google Scholar] [CrossRef]
- Yasmin, S.; Raja, N.I.; Hameed, S.; Brown, J.K. First Association of Pedilanthus leaf curl virus, Papaya leaf curl virus, Cotton leaf curl Kokhran virus, and Papaya leaf curl betasatellite with Symptomatic Chilli Pepper in Pakistan. Plant Dis. 2017, 101, 2155. [Google Scholar] [CrossRef]
- Chen, J.; Zhu, C.; Lin, D.; Sun, Z.-X. The effects of Cd on lipid peroxidation, hydrogen peroxide content and antioxidant enzyme activities in Cd-sensitive mutant rice seedlings. Can. J. Plant Sci. 2007, 87, 49–57. [Google Scholar] [CrossRef]
- Li, Y.; Guo, Q.; Li, Y.; Sun, Y.; Xue, Q.; Lai, H. Streptomyces pactum Act12 controls tomato yellow leaf curl virus disease and alters rhizosphere microbial communities. Biol. Fertil. Soils 2019, 55, 149–169. [Google Scholar] [CrossRef]
- Aebi, H. Catalase in vitro. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 1984; Volume 105, pp. 121–126. [Google Scholar]
- Dickerson, D.; Pascholati, S.; Hagerman, A.E.; Butler, L.; Nicholson, R. Phenylalanine ammonia-lyase and hydroxycinnamate: CoA ligase in maize mesocotyls inoculated with Helminthosporium maydis or Helminthosporium carbonum. Phys. Plant Path. 1984, 25, 111–123. [Google Scholar] [CrossRef]
- Mayer, A.; Harel, E.; Ben-Shaul, R. Assay of catechol oxidase—A critical comparison of methods. Phytochemistry 1966, 5, 783–789. [Google Scholar] [CrossRef]
- Roberts, W.K.; Selitrennikoff, C.P. Plant and Bacterial Chitinases Differ in Antifungal Activity. Microbiology 1988, 134, 169–176. [Google Scholar] [CrossRef]
- Khan, M.F.; Umar, U.U.D. Application of a robust microplate assay to determine induced β-1, 3-glucanase and chitinase activity in the cotton plant. BioTechniques 2021, 70, 202–208. [Google Scholar] [CrossRef]
- Zieslin, N.; Ben Zaken, R. Peroxidase activity and presence of phenolic substances in peduncles of rose flowers. Plant Physiol. Biochem. 1993, 31, 333–339. [Google Scholar]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Ryals, J.A.; Neuenschwander, U.H.; Willits, M.G.; Molina, A.; Steiner, H.-Y.; Hunt, M.D. Systemic acquired resistance. Plant Cell 1996, 8, 1809. [Google Scholar] [CrossRef]
- Pieterse, C.M.; van Loon, L.C. Salicylic acid-independent plant defence pathways. Trends Plant Sci. 1999, 4, 52–58. [Google Scholar] [CrossRef] [PubMed]
- Glazebrook, J. Genes controlling expression of defense responses in Arabidopsis—2001 status. Curr. Opin. Plant Biol. 2001, 4, 301–308. [Google Scholar] [CrossRef]
- Shang, J.; Xi, D.-H.; Xu, F.; Wang, S.-D.; Cao, S.; Xu, M.-Y.; Zhao, P.-P.; Wang, J.-H.; Jia, S.-D.; Zhang, Z.-W.; et al. A broad-spectrum, efficient and nontransgenic approach to control plant viruses by application of salicylic acid and jasmonic acid. Planta 2011, 233, 299–308. [Google Scholar] [CrossRef] [PubMed]
- Faize, M.; Heintz, C.; Cesbron, S.; Chartier, R.; Tharaud, M.; Paulin, J.P.; Brisset, M.N. Induced Resistance to Erwinia amylovora in Apple and Pear. In Proceedings of the IX International Workshop on Fire Blight 590, Hawke’s Bay, New Zealand, 8–12 October 2001; pp. 335–338. [Google Scholar]
- Friedrich, L.; Lawton, K.; Ruess, W.; Masner, P.; Specker, N.; Rella, M.G.; Meier, B.; Dincher, S.; Staub, T.; Uknes, S.; et al. A benzothiadiazole derivative induces systemic acquired resistance in tobacco. Plant J. 1996, 10, 61–70. [Google Scholar] [CrossRef]
- Washington, W.S.; Engleitner, S.; Boontjes, G.; Shanmuganathan, N. Effect of fungicides, seaweed extracts, tea tree oil, and fungal agents on fruit rot and yield in strawberry. Aust. J. Exp. Agric. 1999, 39, 487–494. [Google Scholar] [CrossRef]
- Cui, J.; Bahrami, A.K.; Pringle, E.G.; Hernandez-Guzman, G.; Bender, C.L.; Pierce, N.E.; Ausubel, F.M. Pseudomonas syringae manipulates systemic plant defenses against pathogens and herbivores. Proc. Natl. Acad. Sci. USA 2005, 102, 1791–1796. [Google Scholar] [CrossRef]
- Truman, W.; Bennett, M.H.; Kubigsteltig, I.; Turnbull, C.; Grant, M. Arabidopsis systemic immunity uses conserved defense signaling pathways and is mediated by jasmonates. Proc. Natl. Acad. Sci. USA 2007, 104, 1075–1080. [Google Scholar] [CrossRef] [PubMed]
- Vanitha, S.; Umesha, S. Variations in defense related enzyme activities in tomato during the infection with bacterial wilt pathogen. J. Plant Interact. 2008, 3, 245–253. [Google Scholar] [CrossRef]
- Alscher, R.G.; Erturk, N.; Heath, L.S. Role of superoxide dismutases (SODs) in controlling oxidative stress in plants. J. Exp. Bot. 2002, 53, 1331–1341. [Google Scholar] [CrossRef] [PubMed]
- Hemm, M.R.; Rider, S.D.; Ogas, J.; Murry, D.J.; Chapple, C. Light induces phenylpropanoid metabolism in Arabidopsis roots. Plant J. 2004, 38, 765–778. [Google Scholar] [CrossRef] [PubMed]
- Tahsili, J.; Sharifi, M.; Safaie, N.; Esmaeilzadeh-Bahabadi, S.; Behmanesh, M. Induction of lignans and phenolic compounds in cell culture of Linum album by culture filtrate of Fusarium graminearum. J. Plant Interact. 2014, 9, 412–417. [Google Scholar] [CrossRef]
- Jayaraj, J.; Bhuvaneswari, R.; Rabindran, R.; Muthukrishnan, S.; Velazhahan, R. Oxalic acid-induced resistance to Rhizoctonia solani in rice is associated with induction of phenolics, peroxidase and pathogenesis-related proteins. J. Plant Interact. 2010, 5, 147–157. [Google Scholar] [CrossRef]
- Chandrasekaran, M.; Belachew, S.T.; Yoon, E.; Chun, S.C. Expression of β–1,3-glucanase (GLU) and phenylalanine ammonia-lyase (PAL) genes and their enzymes in tomato plants induced after treatment with Bacillus subtilis CBR05 against Xanthomonas campestris pv. vesicatoria. J. Gen. Plant Pathol. 2016, 83, 7–13. [Google Scholar] [CrossRef]
- Jockusch, H. The Role of Host Genes, Temperature and Polyphenoloxidase in the Necrotization of TMV Infected Tobacco Tissue. J. Phytopathol. 1966, 55, 185–192. [Google Scholar] [CrossRef]
- Mohamed, H.; EL-Hady, A.A.; Mansour, M.; El-Samawaty, A.E.-r. Association of oxidative stress components with resistance to flax powdery mildew. Trop. Plant Pathol. 2012, 37, 386–392. [Google Scholar] [CrossRef]
- Ngadze, E.; Icishahayo, D.; Coutinho, T.A.; van der Waals, J.E.; Insinga, J.K.; Alyokhin, A.; Hao, J.; Ge, T.; Marangoni, N.F.; Baron, A.; et al. Role of Polyphenol Oxidase, Peroxidase, Phenylalanine Ammonia Lyase, Chlorogenic Acid, and Total Soluble Phenols in Resistance of Potatoes to Soft Rot. Plant Dis. 2012, 96, 186–192. [Google Scholar] [CrossRef] [PubMed]
- Singh, H.P.; Kaur, S.; Batish, D.R.; Kohli, R.K. Ferulic acid impairs rhizogenesis and root growth, and alters associated biochemical changes in mung bean (Vigna radiata) hypocotyls. J. Plant Interact. 2014, 9, 267–274. [Google Scholar] [CrossRef]
- Meena, R.K.; Patni, V.; Arora, D. Study on phenolics and their oxidative enzyme in Capsicum annuum L. infected with Geminivirus. Asian J. Exp. Sci. 2008, 22, 307–310. [Google Scholar]
- Yao, K.; De Luca, V.; Brisson, N. Creation of a metabolic sink for tryptophan alters the phenylpropanoid pathway and the susceptibility of potato to Phytophthora infestans. The Plant Cell 1995, 7, 1787–1799. [Google Scholar] [CrossRef]
- Siddique, Z.; Akhtar, K.P.; Hameed, A.; Sarwar, N.; Imran-Ul-Haq; Khan, S.A. Biochemical alterations in leaves of resistant and susceptible cotton genotypes infected systemically by cotton leaf curl Burewala virus. J. Plant Interact. 2014, 9, 702–711. [Google Scholar] [CrossRef]
- Dubery, I.A.; Slater, V. Induced defence responses in cotton leaf disks by elicitors from Verticillium dahliae. Phytochemistry 1997, 44, 1429–1434. [Google Scholar] [CrossRef]
- Abo-Zaid, G.A.; Matar, S.M.; Abdelkhalek, A. Induction of plant resistance against tobacco mosaic virus using the bio-control agent Streptomyces cellulosae isolate Actino 48. Agronomy 2020, 10, 1620. [Google Scholar] [CrossRef]
- Srivastava, S.; Singh, A.K. Changes in catalase activity and total protein content in Urdbean [Vigna mungo (L.) Hepper] plants as a result of UL CV Infection. Indian J. Sci. Res. 2010, 1, 67–69. [Google Scholar]
- Riedle-Bauer, M. Activities of antioxidant enzymes in cucumber plants infected with cucumber mosaic virus. Phyton 1998, 38, 149–157. [Google Scholar]
- Clarke, S.F.; Guy, P.L.; Burritt, D.J.; Jameson, P.E. Changes in the activities of antioxidant enzymes in response to virus infection and hormone treatment. Physiol. Plant. 2002, 114, 157–164. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Ding, Z.; Wu, K.; Yang, L.; Li, Y.; Yang, Z.; Shi, S.; Liu, X.; Zhao, S.; Yang, Z.; et al. Suppression of jasmonic acid-mediated defense by viral-inducible microRNA319 facilitates virus infection in rice. Mol. Plant 2016, 9, 1302–1314. [Google Scholar] [CrossRef]
- Whitham, S.A.; Yang, C.; Goodin, M.M. Global Impact: Elucidating Plant Responses to Viral Infection. Mol. Plant Microbe Interact. 2006, 19, 1207–1215. [Google Scholar] [CrossRef]
- Chen, N.; Hu, M.; Yang, S. The effects of inducing treatments on phenolic metabolism of melon leaves. Acta Horticul Turae Sin. 2010, 37, 1759–1766. [Google Scholar]
- Murphy, A.M.; Chivasa, S.; Singh, D.P.; Carr, J.P. Salicylic acid-induced resistance to viruses and other pathogens: A parting of the ways? Trends Plant Sci. 1999, 4, 155–160. [Google Scholar] [CrossRef]
- Faheed, F.A.; Mahmoud, S.Y. Induction of resistance in Phaseolus vulgaris against TNV by salicylic acid and kinetin. Int. J. Agric. Biol. 2006, 8, 47–51. [Google Scholar]
- Nadia, G.; El-Gamal, G.; Abd-El-Kareem, F.; Fotouh, Y.; El-Mougy, N. Induction of systemic resistance in potato plants against late and early blight diseases using chemical inducers under greenhouse and field conditions. Res. J. Agric. Biol. Sci. 2007, 3, 73–81. [Google Scholar]
- Vernooij, B.; Friedrich, L.; Goy, P.A.; Staub, T.; Kessmann, H.; Ryals, J. 2,6-Dichloroisonicotinic acid-induced resistance to pathogens without the accumulation of salicylic acid. Mol. Plant-Microbe Interact. 1995, 8, 228–234. [Google Scholar] [CrossRef]
- Kauffmann, S.; Legrand, M.; Geoffroy, P.; Fritig, B. Biological function of ‘pathogenesis-related’ proteins: Four PR proteins of tobacco have 1, 3-β-glucanase activity. EMBO J. 1987, 6, 3209–3212. [Google Scholar] [CrossRef]
- Sathiyabama, M.; Manikandan, A. Foliar application of chitosan nanoparticle improves yield, mineral content and boost innate immunity in finger millet plants. Carbohydr. Polym. 2021, 258, 117691. [Google Scholar] [CrossRef] [PubMed]
- Archana, K.; Kaur, S.M.; Pashupat, V.; Javed, A.; Dharmender, P. Role of 2, 6 Dichloroisonicotinic acid inducing resistance in cotton against cotton leaf curl disease. Res. J. Biotechnol. 2020, 15, 5. [Google Scholar]
- Zhang, P.; Vanderschuren, H.; Fütterer, J.; Gruissem, W. Resistance to cassava mosaic disease in transgenic cassava expressing antisense RNAs targeting virus replication genes. Plant Biotechnol. J. 2005, 3, 385–397. [Google Scholar] [CrossRef]
- Ascencio-Ibáñez, J.T.; Sozzani, R.; Lee, T.-J.; Chu, T.-M.; Wolfinger, R.D.; Cella, R.; Hanley-Bowdoin, L. Global analysis of Arabidopsis gene expression uncovers a complex array of changes impacting pathogen response and cell cycle during geminivirus infection. Plant Physiol. 2008, 148, 436–454. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.P.; Moore, C.A.; Gilliland, A.; Carr, J.P. Activation of multiple antiviral defence mechanisms by salicylic acid. Mol. Plant Pathol. 2004, 5, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Lawton, K.; Weymann, K.; Friedrich, L.; Vernooij, B.; Uknes, S.; Ryals, J. Systemic acquired resistance in Arabidopsis re-quires salicylic acid but not ethylene. Mol. Plant Microbe Interact. 1995, 8, 863–870. [Google Scholar] [CrossRef]
- Clarke, J.D.; Volko, S.M.; Ledford, H.; Ausubel, F.M.; Dong, X. Roles of Salicylic Acid, Jasmonic Acid, and Ethylene in cpr-Induced Resistance in Arabidopsis. Plant Cell 2000, 12, 2175–2190. [Google Scholar] [CrossRef]
- Takahashi, H.; Kanayama, Y.; Zheng, M.S.; Kusano, T.; Hase, S.; Ikegami, M.; Shah, J. Antagonistic interactions between the SA and JA signaling pathways in Arabidopsis modulate expression of defense genes and gene-for-gene resistance to cucumber mosaic virus. Plant Cell Physiol. 2004, 45, 803–809. [Google Scholar] [CrossRef]
- Kaloshian, I.; Walling, L.L. Hemipterans as Plant Pathogens. Annu. Rev. Phytopathol. 2005, 43, 491–521. [Google Scholar] [CrossRef] [PubMed]
- Karban, R.; Kuc, J. Induced resistance against pathogens and herbivores: An overview. In Inducible Plant Defenses against Pathogens and Herbivores: Biochemistry, Ecology, and Agriculture; American Phytopathological Society Press: St. Paul, MN, USA, 1999; pp. 1–15. [Google Scholar]
- Zarate, S.I.; Kempema, L.A.; Walling, L.L. Silverleaf Whitefly Induces Salicylic Acid Defenses and Suppresses Effectual Jasmonic Acid Defenses. Plant Physiol. 2006, 143, 866–875. [Google Scholar] [CrossRef]
- Radwan, D.E.M.; Fayez, K.A.; Mahmoud, S.Y.; Hamad, A.; Lu, G. Physiological and metabolic changes of Cucurbita pepo leaves in response to zucchini yellow mosaic virus (ZYMV) infection and salicylic acid treatments. Plant Physiol. Biochem. 2007, 45, 480–489. [Google Scholar] [CrossRef] [PubMed]
- Trejo-Saavedra, D.; García-Neria, M.; Rivera-Bustamante, R. Benzothiadiazole (BTH) induces resistance to Pepper golden mosaic virus (PepGMV) in pepper (Capsicum annuum L.). Biol. Res. 2013, 46, 333–340. [Google Scholar] [CrossRef]
- Thu, Z.; Li, H.-J.; Xie, C.-J.; You, M.-S.; Yang, Z.-M.; Sun, Q.-X.; Liu, Z.-Y. Molecular mapping and chromosomal location of powdery mildew resistance gene in wheat cultivar Tangmai 4. Acta Agron. Sin. 2008, 34, 1193–1198. [Google Scholar]
- Kohler, A.; Schwindling, S.; Conrath, U. Benzothiadiazole-induced priming for potentiated responses to pathogen infection, wounding, and infiltration of water into leaves requires the NPR1/NIM1 gene in Arabidopsis. Plant Physiol. 2002, 128, 1046–1056. [Google Scholar] [CrossRef]
- van Hulten, M.; Pelser, M.; van Loon, L.C.; Pieterse, C.M.J.; Ton, J. Costs and benefits of priming for defense in Arabidopsis. Proc. Natl. Acad. Sci. USA 2006, 103, 5602–5607. [Google Scholar] [CrossRef]
- Conrath, U.; Pieterse, C.M.; Mauch-Mani, B. Priming in plant–pathogen interactions. Trends Plant Sci. 2002, 7, 210–216. [Google Scholar] [CrossRef] [PubMed]












| Enzymatic Activities ± SE | ||||||||
|---|---|---|---|---|---|---|---|---|
| Treatments | SOD | POD | CAT | PPO | PAL | Phenols | β−1,3 Glucanase | Chitinase |
| Control | 451.94 d ± 50.90 | 224.74 f ± 6.57 | 360 e ± 48.39 | 5.66 e ± 0.74 | 66 e ± 3.18 | 50.06 g ± 3.82 | 21.78 d ± 1.13 | 175.09 c ± 9.35 |
| S.A 1 mM * | 795.07 b ± 65.48 | 402.46 b ± 12.64 | 708.9 d ± 31.01 | 11.22 a ± 0.83 | 162 c ± 2.65 | 109.50 c ± 2.87 | 44.53 a ± 2.96 | 207.68 b ± 9.39 |
| S.A 10 Mm | 718.30 bcd ± 96.87 | 295.90 e ± 8.80 | 767.75 c ± 51.70 | 7.75 d ± 0.80 | 158 d ± 4.95 | 97.55 e ± 4.80 | 37.46 b ± 2.29 | 227.97 a ± 14.17 |
| MeJA 1 mM + S.A 1 mM | 630 cd ± 34.21 | 311.26 d ± 8.29 | 707.51 d ± 46.95 | 9.25 b ± 0.24 | 165 b ± 2.96 | 114.22 b ± 3.09 | 37.57 b ± 3.40 | 203.13 b ± 8.58 |
| MeJA 1 mM + S.A 10 mM | 1115.4 a ± 54.61 | 339.45 c ± 14.10 | 894.75 b ± 37.34 | 9.49 b ± 0.51 | 169 a ± 3.40 | 135.97 a ± 4.62 | 34.95 bc ± 3.39 | 213.95 b ± 10.25 |
| BTH 1.1 mM | 818.92 b ± 64.17 | 393.92 b ± 15.26 | 970.7 a ± 59.32 | 8.42 c ± 0.24 | 165 b ± 3.79 | 87.56 f ± 3.64 | 35.77 bc ± 2.78 | 205.25 b ± 10.93 |
| BTH 2.2 mM | 803.86 b ± 37.99 | 447.96 a ± 10.11 | 982 a ± 39.65 | 9.66 b ± 0.27 | 166 ab ± 2.86 | 104.70 d ± 3.99 | 37.53 b ± 2.47 | 208.21 b ± 12.78 |
| MeJA 1 mM+ S.A 5 mM + 1.1 mM BTH | 758.03 bc ± 31.16 | 294.92 e ± 8.23 | 886.53 b ± 34.90 | 9.42 b ± 0.28 | 166 ab ± 4.78 | 111.12 bc ± 5.20 | 33.12 c ± 2.19 | 205.93 b ± 10.60 |
| LSD * | 159.06 | 14.05 | 50.47 | 0.53 | 3.44 | 4.65 | 2.83 | 12.31 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khan, M.F.; Umar, U.U.D.; Alrefaei, A.F.; Rao, M.J. Elicitor-Driven Defense Mechanisms: Shielding Cotton Plants against the Onslaught of Cotton Leaf Curl Multan Virus (CLCuMuV) Disease. Metabolites 2023, 13, 1148. https://doi.org/10.3390/metabo13111148
Khan MF, Umar UUD, Alrefaei AF, Rao MJ. Elicitor-Driven Defense Mechanisms: Shielding Cotton Plants against the Onslaught of Cotton Leaf Curl Multan Virus (CLCuMuV) Disease. Metabolites. 2023; 13(11):1148. https://doi.org/10.3390/metabo13111148
Chicago/Turabian StyleKhan, Muhammad Fahad, Ummad Ud Din Umar, Abdulwahed Fahad Alrefaei, and Muhammad Junaid Rao. 2023. "Elicitor-Driven Defense Mechanisms: Shielding Cotton Plants against the Onslaught of Cotton Leaf Curl Multan Virus (CLCuMuV) Disease" Metabolites 13, no. 11: 1148. https://doi.org/10.3390/metabo13111148
APA StyleKhan, M. F., Umar, U. U. D., Alrefaei, A. F., & Rao, M. J. (2023). Elicitor-Driven Defense Mechanisms: Shielding Cotton Plants against the Onslaught of Cotton Leaf Curl Multan Virus (CLCuMuV) Disease. Metabolites, 13(11), 1148. https://doi.org/10.3390/metabo13111148