Metabolomics-Based Mechanistic Insights into Revealing the Adverse Effects of Pesticides on Plants: An Interactive Review
Abstract
:1. Introduction
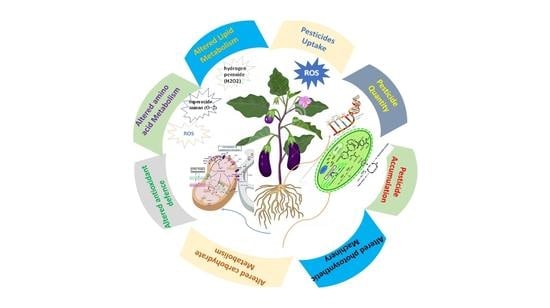
2. Pesticide Toxicity to Agricultural Crops: A Burning Problem
3. Pesticidal Toxicity Mechanisms: An Overview
4. Metabolomics: A Brief Outline
5. Analysis Processes of Metabolomics
Preparation of Samples for Metabolomic Evaluation
| Plant | Organs Used/Involved | Extraction Solvents | Methods of Extraction | Metabolomic Tools/Platform | References |
|---|---|---|---|---|---|
| Chenopodium album (L.) | Leaves, stem | Methanol | Lyophilization + centrifugation | UHPLC-QQQ-MS | [68] |
| Lemna minor (L.) | Leaves | Methanol-ethyl acetate mixture (50:50, v/v)/Ribitol | Extraction, pulverization, sonication | GC/EI/MS | [69] |
| Zea mays (L.) | Root | CH3OH:CHCl3 (2:2, v/v) | - | 1H-HRMAS NMR analysis | [70] |
| Lonicerae japonicae flos | Flower buds | Methanol | Ultrasonication | UPLC/Q-Orbitrap-Full MS | [71] |
| Lactuca sativa (L.) | Leaf tissues | Methanol (80%) and formic acid (0.1%) | Homogenization | UHPLC | [72] |
| Arabidopsis thaliana (L.) | Plant cells | Methanol and H2O | Homogenization | LC/EI/MS | [73] |
| A. thaliana (L.) | Leaves | Methanol, chloroform, and H2O (chilled) | Homogenization | LDI/MS | [74] |
| Cucumis sativus (L.) | Leaves | Methanol/water (100:0), acetonitrile/water (80:20) acetone/water | Homogenization+ centrifugation | LS–MS/MS | [75] |
| Solanum lycopersicum (L.) | Leaves | Methanol (70%) | Centrifugation | UHPLC/Q-TOF | [76] |
| Solanum tuberosum (L.) | Tubers | Methanol | Homogenization | UPLC-IMS-QtoF | [77] |
| Oryza sativa (L.) | Leaves and seeds | Acetonitrile/isopropanol/water (3/3/2, v/v/v) | Centrifugation | GC–MS | [78] |
| O. sativa (L.) | Leaves | Methanol | Solvent extraction + homogenization | GC–MS | [79] |
| S. lycopersicum (L.) | Fruit | Acetonitrile/acetic acid/anhydrous MgSO4/sodium acetate | Solvent extraction | LC–MS | [80] |
| O. sativa (L.) | Leaves | Methanol and H2O | Solvent extraction + derivatization/homogenization | HS-SPME/GC-MS | [81] |
| C. sativus (L.) | Fruit | Methanol/ H2O and chloroform | Extraction/homogenization | UHPLC-Q-Orbitrap-HRMS | [82] |
| O. sativa (L.) | Leaves | Methanol/methyl-tertiary butyl ether/H2O | Solvent extraction/homogenization | LC–MS | [83] |
| Tasmannia piperita (L.) | Leaves | Methanol | Solvent extraction | UHPLC-HRMS | [84] |
| Vitis vinifera (L.) | Leaves | Methanol/chloroform/H2O | Solvent extraction/fractionation/homogenization | FTICR–MS | [64] |
| O. sativa (L.) | Leaves | Acetonitrile/ isopropanol/water (3:3:2, v/v/v) | Solvent extraction + ultrasonication | GC–MS | [85] |
| Glycine max merr | Leaves | Methanol/acetonitrile/deionized water, 2/2/1, v/v/v | Ultrasonication/ centrifugation | LC–MS/MS | [86] |
| Helianthus annuus | Plants | Perchloric acid | - | 1D 1H NMR | [87] |
| Brassica oleracea | Leaves | Methanol/chloroform/water in a 2:2:1 | Solvent extraction+ centrifugation | NMR | [88] |
| L. sativa (L.) | Leaves | Acetone: hexane (1:1, v/v) | Solvent extraction | GC×GC–MS | [89] |
| Beta vulgaris (L.) | Roots | Ethanol | Solvent extraction/ homogenization | UPLC Q-TOF LC-MS | [90] |
6. Metabolomics Approaches Used to Assess Pesticide–Plant Interactions
6.1. Nuclear Magnetic Resonance (NMR)-Based Metabolomics
6.2. MS-Based Metabolomics
6.2.1. Gas Chromatography–Mass Spectrophotometry (GC–MS)-Based Metabolomics
6.2.2. Liquid Chromatography–Mass Spectrophotometry (LC–MS)-Based Metabolomics
6.2.3. Gas Chromatography/Electron Impact Mass Spectrometry (GC/EI/MS)-Based Metabolomics
6.2.4. Gas Chromatography–Time-of-Flight Mass Spectrometry (GC–TOF MS)-Based Metabolomics
6.2.5. Combined NMR- and MS-Based Metabolomics
6.2.6. Ultra-High-Performance Liquid Chromatography (UHPLC)-Based Metabolomics
6.2.7. Ultra-High-Performance Liquid Chromatography Coupled with High-Resolution Mass Spectrometry (UHPLC–HRMS)
6.2.8. Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry (MALDI–TOF MS)-Based Metabolomics
6.2.9. Ultra-Performance Liquid Chromatography–Ion Mobility Spectroscopy–Quadrupole Time-of-Flight Mass Spectrometry (UPLC–IMS–QtoF)-Based Metabolomics
| Pesticide Used | Chemical Class/Family | Dose Rate | Crop/Vegetable Used | Organ Involved | Way of Analyses/ Platform | Observation | Changes Observed | References |
|---|---|---|---|---|---|---|---|---|
| Mancozeb | Dithiocarbamate family | 2.0 mg/L | Lectuca sativa (Lettuce) | Leaves | NMR-HRMAS | Negatively affected |
| [33] |
| Imidacloprid (IMD) and fenvalerate (FVE) | IMD; neonicotinoids, FVE; pyrethroid | 10 mg/L | Lactuca sativa L. (lettuce) | Leaves | UHPLC | Negative effect |
| [123] |
| Chlorpyrifos | Chlorinated organophosphate | 0.576, 0.720, and 1.080 kg a. i./ha | Oryza sativa L (rice) | Leaves | GC-MS | Negatively affected |
| [134] |
| Isoprocarb, carbofuran, and carbaryl | Carbamates | 5.0 μg mL−1 | Brassica campestris L. ssp. Chinensis (mustard) and Makino var. communis | Leaves | SPME | Negatively affected |
| [135] |
| Chlorpyrifos | Chlorinated organophosphate | 0.02%, 0.06%, and 0.08% | Phaseolus vulgaris L. (Common bean) | Pod and beans | LC–MS and MALDI–TOF MS | Negatively affected |
| [125] |
| Acetamiprid (ACE) and cyromazine (CYR) | ACD; chloropyridinyl neonicotinoids, CYMZ; aminotriazines | 540 g a.i. ha 150 g a.i. ha | Vigna unguiculata L. (cowpea) | Leaves | LC–MS/MS | Negatively affected |
| [105] |
| Nano copper pesticides | 400–800 mg/kg | Cucumis sativus L. (cucumber) | LC–MS/MS |
| [118] | |||
| Glyphosate (GP) and metribuzin (MBN) | GP; organophosphorus, MBN; triazinones | 0.5, 1.0, 5.0, 10, 25 and 50 ppm | Lemna minor L. | Leaves | GC/EI/MS | Negative effect |
| [69] |
| Lindane (HCH) and chlordecone (CLD) | LCH; organochlorine, CLD; organochlorine | 2.5 µM to 25 µM | Zea mays L. (maize) | Root tips | 1H-HR-MAS NMR | Negative effect |
| [70] |
| Fungicides | - | - | Solanum tuberosum L. (potato) | Tubers | UPLC–IMS–QtoF | Negative effect |
| [77] |
| Butachlor (BUTA), chlorpyrifos (CPF), tricyclazole (TZL) | BUTA; acetanilide class, CPF; organophosphates, TZL; triazolobenzothiazoles | CPF = 0.576, 0.720 and 1.080 kg a.i./ha. BUT = 0.90 and 1.574, 2.624 kg a.i./h. | Oryza sativa L. (rice) | Leaves | GC-MS | Negative effect |
| [78] |
| Chlorpyrifos | Organophosphate | 2.0, 5.0 and 20.0 mg L−1 | Oryza sativa L. (Rice) | Leaves and roots | LC–QTOF/MS | Negatively affected |
| [136] |
| Thiamathoxam | Neonicotinoid | 500 mgL−1 | Camellia sinensis L. | Leaves | GC–MS and HPLC | Negatively affected |
| [137] |
| Gulfosinate | - | 1, 5, 10, and 15% v/v | Stenotaphrum secundatum L. | Plant | GC–MS | Negatively affected |
| [138] |
| Imazamox | Imidazolinone | 0.036, 0.035, and 0.203 mg/L | Lemna minor L. | Leaves | LC–MS | Negatively affected |
| [117] |
| Perfluorooctanesulfonic acid | - | 0, 25, and 50 mg/kg | Triticum aestivum L. | Roots and grains | HPLC/MS/MS | Negatively affected |
| [139] |
| NMR | MS | |
|---|---|---|
| Sensitivity | Low but can be enhanced using cryo- and microprobes, dynamic nuclear polarisation, and greater field strengths. | High with a nanomolar detection threshold |
| Selectivity | Despite the fact that there are only a few selective experiments available, such as selective TOCSY, it is typically employed for nonselective analysis. | Can be used for both targeted and untargeted (selected and non-targeted) studies |
| Sample measurement | One measurement allows for the detection of all metabolites with an NMR concentration level. | For various kinds of metabolites, different chromatographic methods are typically required. |
| Sample recovery | Numerous analyses can be performed on the same sample without causing any damage; the sample can be recovered and kept for a long time. | Destructive method, but requires a small sample size |
| Reproducibility | Very high | Moderate |
| Number of detectable metabolites | 30–100 | 300–1000+ (depending on whether GC–MS or LC–MS is used) |
| Sample preparation | Little sample preparation is required | More difficult; requires different columns and ionisation condition optimization |
| Tissue samples | Yes, using HR-MAS NMR tissue samples analysed directly | No, requires tissue extraction |
| Target analysis | Inapplicable to targeted analysis | More effective for specialised analysis |
| Sample analysis time | Quick—one measurement can be used to analyse the entire sample. | Longer and uses various chromatography methods depending on the metabolites being examined |
| In vivo studies | Yes—widely used for 1H magnetic resonance spectroscopy (and to a lesser degree 31P and 13C) | No—although desorption electrospray ionization (DESI) may be a useful way to sample tissues in a minimally invasive way during surgery |
| Instrument cost | More expensive and occupies more space | More affordable (cheaper) and compact |
| Sample cost | Low cost per sample | High cost per sample |
7. Challenges in Metabolomics and Future Prospects
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| GC–MS | Gas chromatography–mass spectrophotometry |
| LC–MS | Liquid chromatography–mass spectrophotometry |
| HPLC | High-performance liquid chromatography |
| UPLC–IMS–QtoF | Ultra-performance liquid chromatography–ion mobility spectroscopy–quadrupole time-of-flight mass spectrometry |
| MALDI–TOF MS | Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry |
| GC/EI/MS | Gas chromatography/electron impact mass spectrometry |
| 1H-HR-MAS NMR | High-resolution magic angle-spinning nuclear magnetic resonance |
| ROS | Reactive oxygen species |
| H2O2 | Hydrogen peroxide |
| (O−2) | Superoxide anions |
| RBOH1 | Respiratory burst oxidase homologous 1 |
| NAD | Nicotinamide adenine dinucleotide |
| NADP | Nicotinamide adenine dinucleotide phosphate |
| ATP | Adenosine triphosphate |
| (ΔΨm) | Mitochondrial membrane potential |
| DNA | Deoxyribonucleic acid |
| TCA | Tricarboxylic acid cycle |
| GC-LRMS | Gas chromatography coupled to low-resolution mass spectrometry |
| NMR | Nuclear magnetic resonance |
| DI–FTICR–MS | Direct-infusion Fourier-transform ion cyclotron-resonance |
| DMSO | Dimethyl sulfoxide |
| HMDS | Hexamethyldisilazane |
| MTBSTFA | N-tert-butyldimethylsilyl-N-methyltrifluoroacetamide |
| FTICR–MS | Fourier-transform ion cyclotron resonance (FTICR) mass spectrometry |
| GC–TOF MS | Gas chromatography (GC) coupled to time-of-flight mass spectrometry |
| ICP–MS | Inductively coupled plasma mass spectrometry |
| UHPLC | Ultra-high-performance liquid chromatography |
| 2D-DIGE | Two-dimensional difference gel electrophoresis |
| QuEChERS | Quick, easy, cheap, effective, rugged, and safe |
| MS (UPLC–TWIMS–QTOF) | Ultra-performance liquid chromatography system coupled to a high-resolution quadrupole/traveling wave ion mobility spectrometry/time-of-flight |
References
- USEPA—United States of Environmental Protection Agengy. About Pesticides. U.S. EPA. Available online: https://www.epa.gov/pesticide-registration/about-pesticide-registration (accessed on 27 August 2014).
- Mesnage, R.; Székács, A.; Zaller, J.G. Herbicides: Brief history, agricultural use, and potential alternatives for weed control. In Herbicides; Chemistry, Efficacy, Toxicology, and Environmental Impacts Emerging Issues in Analytical Chemistry; Elsevier: Amsterdam, The Netherlands, 2021; pp. 1–20. [Google Scholar]
- Wojciechowska, M.; Stepnowski, P.; Gołębiowski, M. The use of insecticides to control insect pests. Invertebr. Surviv. J. 2016, 13, 210–220. [Google Scholar]
- Singh, S.; Kumar, V.; Dhanjal, D.S.; Singh, J. Herbicides and plant growth regulators: Current developments and future challenges. In Natural Bioactive Products in Sustainable Agriculture; Springer: Singapore, 2020; pp. 67–81. [Google Scholar]
- Price, C.L.; Parker, J.E.; Warrilow, A.G.; Kelly, D.E.; Kelly, S.L. Azole fungicides–understanding resistance mechanisms in agricultural fungal pathogens. Pest Manag. Sci. 2015, 71, 1054–1058. [Google Scholar] [CrossRef]
- Shahid, M.; Khan, M.S. Ecotoxicological implications of residual pesticides to beneficial soil bacteria: A review. Pestic. Biochem. Physiol. 2022, 188, 105272. [Google Scholar] [CrossRef]
- Yadav, I.C.; Devi, N.L.; Syed, J.H.; Cheng, Z.; Li, J.; Zhang, G.; Jones, K.C. Current status of persistent organic pesticides residues in air, water, and soil, and their possible effect on neighboring countries: A comprehensive review of India. Sci. Total Environ. 2015, 511, 123–137. [Google Scholar] [CrossRef]
- Shahid, M.; Khan, M.S.; Ahmed, B.; Syed, A.; Bahkali, A.H. Physiological disruption, structural deformation and low grain yield induced by neonicotinoid insecticides in chickpea: A long term phytotoxicity investigation. Chemosphere 2021, 262, 128388. [Google Scholar] [CrossRef] [PubMed]
- Shahid, M.; Khan, M.S.; Syed, A.; Marraiki, N.; Elgorban, A.M. Mesorhizobium ciceri as biological tool for improving physiological, biochemical and antioxidant state of Cicer aritienum (L.) under fungicide stress. Sci. Rep. 2021, 11, 1–18. [Google Scholar] [CrossRef]
- Shahid, M.; Manoharadas, S.; Chakdar, H.; Alrefaei, A.F.; Albeshr, M.F.; Almutairi, M.H. Biological toxicity assessment of carbamate pesticides using bacterial and plant bioassays: An in-vitro approach. Chemosphere 2021, 278, 130372. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.; Shahid, M.; Khan, M.S.; Syed, A.; Bahkali, A.H.; Elgorban, A.M.; Pichtel, J. Fungicide-tolerant plant growth-promoting rhizobacteria mitigate physiological disruption of white radish caused by fungicides used in the field cultivation. Int. J. Environ. Res. Public Health 2020, 17, 7251. [Google Scholar] [CrossRef] [PubMed]
- Shahid, M.; Khan, M.S.; Kumar, M. Kitazin-pea interaction: Understanding the fungicide induced nodule alteration, cytotoxicity, oxidative damage and toxicity alleviation by Rhizobium leguminosarum. RSC Adv. 2019, 9, 16929–16947. [Google Scholar] [CrossRef]
- Shahid, M.; Khan, M.S. Fungicide tolerant Bradyrhizobium japonicum mitigate toxicity and enhance greengram production under hexaconazole stress. J. Environ. Sci. 2019, 78, 92–108. [Google Scholar] [CrossRef]
- Shahid, M.; Ahmed, B.; Zaidi, A.; Khan, M.S. Toxicity of fungicides to Pisum sativum: A study of oxidative damage, growth suppression, cellular death and morpho-anatomical changes. RSC Adv. 2018, 8, 38483–38498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, X.; Wang, N.; Sun, S.; Geng, L.; Guo, N.; Liu, A.; Chen, S.; Ahammed, G.J. Reactive oxygen species signalling is involved in melatonin-induced reduction of chlorothalonil residue in tomato leaves. J. Hazard. Mater. 2023, 443, 130212. [Google Scholar] [CrossRef] [PubMed]
- Lydon, J.; Duke, S.O. Pesticide effects on secondary metabolism of higher plants. Pestic. Sci. 1989, 25, 361–373. [Google Scholar] [CrossRef]
- Li, X.; Li, Y.; Ahammed, G.J.; Zhang, X.N.; Ying, L.; Zhang, L.; Yan, P.; Zhang, L.P.; Li, Q.Y.; Han, W.Y. RBOH1-dependent apoplastic H2O2 mediates epigallocatechin-3-gallate-induced abiotic stress tolerance in Solanum lycopersicum L. Environ. Exp. Bot. 2019, 161, 357–366. [Google Scholar] [CrossRef]
- Shahid, M.; Khan, M. Glyphosate induced toxicity to chickpea plants and stress alleviation by herbicide tolerant phosphate solubilizing Burkholderia cepacia PSBB1 carrying multifarious plant growth promoting activities. 3 Biotech 2018, 8, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.M.; Esser, L.; Zhou, F.; Li, C.; Zhou, Y.H.; Yu, C.A.; Qin, Z.H.; Xia, D. Studies on inhibition of respiratory cytochrome bc 1 complex by the fungicide pyrimorph suggest a novel inhibitory mechanism. PLoS ONE 2014, 9, e93765. [Google Scholar]
- Shahid, M.; Khan, M.S.; Zaidi, A. Fungicide toxicity to legumes and its microbial remediation: A current perspective. Pestic. Crop. Prod. Physiol. Biochem. Action 2020, 15–33. [Google Scholar] [CrossRef]
- Sharma, I.; Bhardwaj, R.; Pati, P.K. Exogenous application of 28-homobrassinolide modulates the dynamics of salt and pesticides induced stress responses in an elite rice variety Pusa Basmati-1. J. Plant Growth Reg. 2015, 34, 509–518. [Google Scholar] [CrossRef]
- Shakir, S.K.; Irfan, S.; Akhtar, B.; Daud, M.K.; Taimur, N.; Azizullah, A. Pesticide-induced oxidative stress and antioxidant responses in tomato (Solanum lycopersicum) seedlings. Ecotoxicology 2018, 27, 919–935. [Google Scholar] [CrossRef]
- Chen, S.; Yin, C.; Strasser, R.J.; Yang, C.; Qiang, S. Reactive oxygen species from chloroplasts contribute to 3-acetyl-5-isopropyltetramic acid-induced leaf necrosis of Arabidopsis thaliana. Plant Physiol. Biochem. 2012, 52, 38–51. [Google Scholar] [CrossRef]
- Stanley, J.; Preetha, G.; Stanley, J. Pesticide Toxicity to Non-Target Organisms; Springer: Berlin/Heidelberg, Germany, 2016. [Google Scholar]
- Hall, R.D. Plant metabolomics: From holistic hope, to hype, to hot topic. New Phytol. 2006, 169, 453–468. [Google Scholar] [CrossRef]
- Rinschen, M.M.; Ivanisevic, J.; Giera, M.; Siuzdak, G. Identification of bioactive metabolites using activity metabolomics. Nat. Rev. Mol. Cell Biol. 2019, 20, 353–367. [Google Scholar] [CrossRef]
- Allwood, J.W.; De Vos, R.C.; Moing, A.; Deborde, C.; Erban, A.; Kopka, J.; Goodacre, R.; Hall, R.D. Plant metabolomics and its potential for systems biology research: Background concepts, technology, and methodology. Methods Enzymol. 2011, 500, 299–336. [Google Scholar]
- Wishart, D.S. Metabolomics for investigating physiological and pathophysiological processes. Physiol. Rev. 2019, 99, 1819–1875. [Google Scholar] [CrossRef] [PubMed]
- Viant, M.R. Recent developments in environmental metabolomics. Mol. Biosyst. 2008, 4, 980–986. [Google Scholar] [CrossRef] [PubMed]
- Matich, E.K.; Soria, N.G.C.; Aga, D.S.; Atilla-Gokcumen, G.E. Applications of metabolomics in assessing ecological effects of emerging contaminants and pollutants on plants. J. Hazard. Mater. 2019, 373, 527–535. [Google Scholar] [CrossRef]
- Dreher, K. Putting the plant metabolic network pathway databases to work: Going offline to gain new capabilities. In Plant Metabolism; Humana Press: Totowa, NJ, USA, 2014; pp. 151–171. [Google Scholar]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Pereira, S.I.; Figueiredo, P.I.; Barros, A.S.; Dias, M.C.; Santos, C.; Duarte, I.F.; Gil, A.M. Changes in the metabolome of lettuce leaves due to exposure to mancozeb pesticide. Food Chem. 2014, 154, 291–298. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Liu, J.; Wen, R.; Chen, Q.; Kong, B. Metabolomics profiling reveals defense strategies of Pediococcus pentosaceus R1 isolated from Harbin dry sausages under oxidative stress. LWT 2021, 135, 110041. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhang, L.; Zhao, C.; Hu, C.; Li, Y.; Zhao, J.; Zhang, J.; Li, L.; Chang, Y.; Wang, F.; et al. Metabolic responses of rice leaves and seeds under transgenic backcross breeding and pesticide stress by pseudo-targeted metabolomics. Metabolomics 2015, 11, 1802–1814. [Google Scholar] [CrossRef]
- Hu, L.; Liu, J.; Zhang, W.; Wang, T.; Zhang, N.; Lee, Y.H.; Lu, H. Functional metabolomics decipher biochemical functions and associated mechanisms underlie small-molecule metabolism. Mass Spectrom. Rev. 2020, 39, 417–433. [Google Scholar] [CrossRef] [PubMed]
- Labine, L.M.; Simpson, M.J. The use of nuclear magnetic resonance (NMR) and mass spectrometry (MS)–based metabolomics in environmental exposure assessment. Curr. Opinion Environ. Sci. Health 2020, 15, 7–15. [Google Scholar] [CrossRef]
- Emwas, A.H.; Roy, R.; McKay, R.T.; Tenori, L.; Saccenti, E.; Gowda, G.N.; Raftery, D.; Alahmari, F.; Jaremko, L.; Jaremko, M.; et al. NMR spectroscopy for metabolomics research. Metabolites 2019, 9, 123. [Google Scholar] [CrossRef] [Green Version]
- Bothwell, J.H.; Griffin, J.L. An introduction to biological nuclear magnetic resonance spectroscopy. Biol. Rev. 2011, 86, 493–510. [Google Scholar] [CrossRef]
- Shaykhutdinov, R.A.; MacInnis, G.D.; Dowlatabadi, R.; Weljie, A.M.; Vogel, H.J. Quantitative analysis of metabolite concentrations in human urine samples using 13C {1H} NMR spectroscopy. Metabolomics 2009, 5, 307–317. [Google Scholar] [CrossRef]
- Traverso, A.; Wee, L.; Dekker, A.; Gillies, R. Repeatability and reproducibility of radiomic features: A systematic review. Int. J. Radiation Oncol. Biol. Phys. 2018, 102, 1143–1158. [Google Scholar] [CrossRef]
- Valentino, G.; Graziani, V.; D’Abrosca, B.; Pacifico, S.; Fiorentino, A.; Scognamiglio, M. NMR-based plant metabolomics in nutraceutical research: An overview. Molecules 2020, 25, 1444. [Google Scholar] [CrossRef]
- Finehout, E.J.; Lee, K.H. An introduction to mass spectrometry applications in biological research. Biochem. Mol. Biol. Educ. 2004, 32, 93–100. [Google Scholar] [CrossRef]
- Blakley, C.R.; Carmody, J.J.; Vestal, M.L. Liquid chromatograph-mass spectrometer for analysis of non-volatile samples. Analytic. Chem. 1980, 52, 1636–1641. [Google Scholar] [CrossRef]
- Zhao, S.; Li, L. Chemical derivatization in LC-MS-based metabolomics study. Trends Analytic. Chem. 2020, 131, 115988. [Google Scholar] [CrossRef]
- Want, E.J.; Cravatt, B.F.; Siuzdak, G. The expanding role of mass spectrometry in metabolite profiling and characterization. Chembiochem 2005, 6, 1941–1951. [Google Scholar] [CrossRef] [PubMed]
- Cajka, T.; Fiehn, O. Toward merging untargeted and targeted methods in mass spectrometry-based metabolomics and lipidomics. Anal. Chem. 2016, 88, 524–545. [Google Scholar] [CrossRef]
- Hill, C.B.; Roessner, U. Metabolic profiling of plants by GC–MS. In Handbook of Plant Metabolomics; Wiley-VCH Verlag GmbH & Co. KGaA: Weinheim, Germany, 2013; pp. 1–23. [Google Scholar] [CrossRef]
- Terracio, L.; Schwabe, K.G. Freezing and drying of biological tissues for electron microscopy. J. Histochem. Cytochem. 1981, 29, 1021–1028. [Google Scholar] [CrossRef]
- Miazek, K.; Kratky, L.; Sulc, R.; Jirout, T.; Aguedo, M.; Richel, A.; Goffin, D. Effect of organic solvents on microalgae growth, metabolism and industrial bioproduct extraction: A review. Int. J. Mol. Sci. 2017, 18, 1429. [Google Scholar] [CrossRef]
- Joshi, D.R.; Adhikari, N. An overview on common organic solvents and their toxicity. J. Pharm. Res. Int. 2019, 28, 1–18. [Google Scholar] [CrossRef]
- Nelson, W.M. Green Solvents for Chemistry: Perspectives and Practice; Oxford University Press: Oxford, UK, 2003. [Google Scholar]
- Nanda, B.; Sailaja, M.; Mohapatra, P.; Pradhan, R.K.; Nanda, B.B. Green solvents: A suitable alternative for sustainable chemistry. Mater. Today Proc. 2021, 47, 1234–1240. [Google Scholar] [CrossRef]
- Zalewska, K.A.M. Development of Novel Ionic Liquids Based on Biological Molecules. Ph.D. Thesis, Universidade NOVA de Lisboa, Lisbon, Portugal, 2014. [Google Scholar]
- Yang, Z.; Pan, W. Ionic liquids: Green solvents for nonaqueous bio catalysis. Enzym. Microb. Technol. 2005, 37, 19–28. [Google Scholar] [CrossRef]
- Ramos, L.; Kristenson, E.M.; Brinkman, U.T. Current use of pressurised liquid extraction and subcritical water extraction in environmental analysis. J. Chromatogr. A 2002, 975, 3–29. [Google Scholar] [CrossRef] [PubMed]
- Sapkale, G.N.; Patil, S.M.; Surwase, U.S.; Bhatbhage, P.K. Supercritical fluid extraction. Int. J. Chem. Sci. 2010, 8, 729–743. [Google Scholar]
- Abbasi, N.M.; Farooq, M.Q.; Anderson, J.L. Modulating solvation interactions of deep eutectic solvents formed by ammonium salts and carboxylic acids through varying the molar ratio of hydrogen bond donor and acceptor. J. Chromatogr. A 2021, 1643, 462011. [Google Scholar] [CrossRef]
- Lam, S.M.; Tian, H.; Shui, G. Lipidomics, en route to accurate quantitation. Biochim. Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 2017, 1862, 752–761. [Google Scholar] [CrossRef]
- Salem, M.A.; Jüppner, J.; Bajdzienko, K.; Giavalisco, P. Protocol: A fast, comprehensive and reproducible one-step extraction method for the rapid preparation of polar and semi-polar metabolites, lipids, proteins, starch and cell wall polymers from a single sample. Plant Methods 2016, 12, 1–15. [Google Scholar] [CrossRef]
- Corrêa, P.S.; Morais Júnior, W.G.; Martins, A.A.; Caetano, N.S.; Mata, T.M. Microalgae biomolecules: Extraction, separation and purification methods. Processes 2020, 9, 10. [Google Scholar] [CrossRef]
- Lee, J.H.; Zhu, J. Analyses of short-chain fatty acids and exhaled breath volatiles in dietary intervention trials for metabolic diseases. Exp. Biol. Med. 2021, 246, 778–789. [Google Scholar] [CrossRef] [PubMed]
- Smart, K.F.; Aggio, R.B.; Van Houtte, J.R.; Villas-Bôas, S.G. Analytical platform for metabolome analysis of microbial cells using methyl chloroformate derivatization followed by gas chromatography–mass spectrometry. Nat. Protoc. 2010, 5, 1709–1729. [Google Scholar] [CrossRef]
- Maia, M.; Monteiro, F.; Sebastiana, M.; Marques, A.P.; Ferreira, A.E.; Freire, A.P.; Cordeiro, C.; Figueiredo, A.; Silva, M.S. Metabolite extraction for high-throughput FTICR-MS-based metabolomics of grapevine leaves. EuPA Open Proteom. 2016, 12, 4–9. [Google Scholar] [CrossRef] [PubMed]
- Brown, S.A.; Simpson, A.J.; Simpson, M.J. Evaluation of sample preparation methods for nuclear magnetic resonance metabolic profiling studies with Eisenia fetida. Environ. Toxicol. Chem. Int. J. 2008, 27, 828–836. [Google Scholar] [CrossRef]
- Guy, G.R.; Philip, R.; Tan, Y.H. Analysis of cellular phosphoproteins by two-dimensional gel electrophoresis: Applications for cell signaling in normal and cancer cells. Electrophoresis 1994, 15, 417–440. [Google Scholar] [CrossRef] [PubMed]
- McKelvie, J.R.; Yuk, J.; Xu, Y.; Simpson, A.J.; Simpson, M.J. 1H NMR and GC/MS metabolomics of earthworm responses to sub-lethal DDT and endosulfan exposure. Metabolomics 2009, 5, 84–94. [Google Scholar] [CrossRef]
- Liu, N.; An, X.; Wang, Y.; Qi, J. Metabolomics Analysis Reveals the Effect of Fermentation to Secondary Metabolites of Chenopodium album L. Based on UHPLC-QQQ-MS. Fermentation 2023, 9, 100. [Google Scholar] [CrossRef]
- Kostopoulou, S.; Ntatsi, G.; Arapis, G.; Aliferis, K.A. Assessment of the effects of metribuzin, glyphosate, and their mixtures on the metabolism of the model plant Lemna minor L. applying metabolomics. Chemosphere 2020, 239, 124582. [Google Scholar] [CrossRef] [PubMed]
- Blondel, C.; Khelalfa, F.; Reynaud, S.; Fauvelle, F.; Raveton, M. Effect of organochlorine pesticides exposure on the maize root metabolome assessed using high-resolution magic-angle spinning 1H-NMR spectroscopy. Environ. Pollut. 2016, 214, 539–548. [Google Scholar] [CrossRef] [PubMed]
- Hui-Qin, P.A.N.; Heng, Z.H.O.U.; Shui, M.I.A.O.; De-An, G.U.O.; Zhang, X.L.; Qing, H.U.; Xiu-Hong, M.A.O.; Shen, J.I. Plant metabolomics for studying the effect of two insecticides on comprehensive constituents of Lonicerae Japonicae Flos. Chin. J. Nat. Med. 2021, 19, 70–80. [Google Scholar]
- Assefa, A.D.; Hur, O.S.; Hahn, B.S.; Kim, B.; Ro, N.Y.; Rhee, J.H. Nutritional metabolites of red pigmented lettuce (Lactuca sativa) germplasm and correlations with selected phenotypic characters. Foods 2021, 10, 2504. [Google Scholar] [CrossRef] [PubMed]
- Sarry, J.E.; Kuhn, L.; Ducruix, C.; Lafaye, A.; Junot, C.; Hugouvieux, V.; Jourdain, A.; Bastien, O.; Fievet, J.B.; Vailhen, D.; et al. The early responses of Arabidopsis thaliana cells to cadmium exposure explored by protein and metabolite profiling analyses. Proteomics 2006, 6, 2180–2198. [Google Scholar] [CrossRef]
- Zhou, J.; Wang, J.; Shi, K.; Xia, X.J.; Zhou, Y.H.; Yu, J.Q. Hydrogen peroxide is involved in the cold acclimation-induced chilling tolerance of tomato plants. Plant Physiol. Biochem. 2012, 60, 141–149. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Adeleye, A.S.; Zhao, L.; Minakova, A.S.; Anumol, T.; Keller, A.A. Antioxidant response of cucumber (Cucumis sativus) exposed to nano copper pesticide: Quantitative determination via LC-MS/MS. Food Chem. 2019, 270, 47–52. [Google Scholar] [CrossRef]
- Pretali, L.; Bernardo, L.; Butterfield, T.S.; Trevisan, M.; Lucini, L. Botanical and biological pesticides elicit a similar induced systemic response in tomato (Solanum lycopersicum) secondary metabolism. Phytochemistry 2016, 130, 56–63. [Google Scholar] [CrossRef]
- Claassen, C.; Ebel, E.; Kuballa, J.; Rohn, S. Impacts of fungicide treatment and conventional fertilization management on the potato metabolome (Solanum tuberosum L.) evaluated with UPLC-IMS-QToF. J. Agric Food Chem. 2019, 67, 11542–11552. [Google Scholar] [CrossRef]
- Liu, N.; Zhu, L. Metabolomic and transcriptomic investigation of metabolic perturbations in Oryza sativa L. triggered by three pesticides. Environ. Sci. Tech. 2020, 54, 6115–6124. [Google Scholar] [CrossRef]
- Zhou, J.; Zhang, L.; Chang, Y.; Lu, X.; Zhu, Z.; Xu, G. Alteration of leaf metabolism in Bt-transgenic rice (Oryza sativa L.) and its wild type under insecticide stress. J. Proteome Res. 2012, 11, 4351–4360. [Google Scholar] [CrossRef]
- Siano, G.G.; Pérez, I.S.; García, M.D.G.; Galera, M.M.; Goicoechea, H.C. Multivariate curve resolution modeling of liquid chromatography–mass spectrometry data in a comparative study of the different endogenous metabolites behavior in two tomato cultivars treated with carbofuran pesticide. Talanta 2011, 85, 264–275. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Xi, J.; Xu, D.; Jin, Y.; Wu, F.; Tong, Q.; Yin, Y.; Xu, X. A comparative HS-SPME/GC-MS-based metabolomics approach for discriminating selected japonica rice varieties from different regions of China in raw and cooked form. Food Chem. 2022, 385, 132701. [Google Scholar] [CrossRef] [PubMed]
- Pan, L.; Zhou, C.; Jing, J.; Zhuang, M.; Zhang, J.; Wang, K.; Zhang, H. Metabolomics analysis of cucumber fruit in response to foliar fertilizer and pesticides using UHPLC-Q-Orbitrap-HRMS. Food Chem. 2022, 369, 130960. [Google Scholar] [CrossRef] [PubMed]
- Mahdavi, V.; Farimani, M.M.; Fathi, F.; Ghassempour, A. A targeted metabolomics approach toward understanding metabolic variations in rice under pesticide stress. Anal. Biochem. 2015, 478, 65–72. [Google Scholar] [CrossRef] [PubMed]
- Amoroso, V.B.; Mendez, R.A.; Junio, H.A.; Molino, R.J.E.J.; Pescadero, I.R.; Villalobos, A.P. Characterization of a natural fungicide from an indigenous plant Tasmannia piperita (Hook. f.) Miers Extract: Stability, Metabolomics, and In silico Studies. Philipp. J. Sci. 2021, 150, 355–370. [Google Scholar] [CrossRef]
- Das, A.B.; Goud, V.V.; Das, C. Extraction of phenolic compounds and anthocyanin from black and purple rice bran (Oryza sativa L.) using ultrasound: A comparative analysis and phytochemical profiling. Indus. Crops Prod. 2017, 95, 332–341. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, Q.; Yu, Y.; Li, X.; Tan, H. Integrated proteomics, metabolomics and physiological analyses for dissecting the toxic effects of halo-sulfuron-methyl on soybean seedlings (Glycine max merr.). Plant Physiol. Biochem. 2020, 157, 303–315. [Google Scholar] [CrossRef] [PubMed]
- Kruger, N.J.; Troncoso-Ponce, M.A.; Ratcliffe, R.G. 1H NMR metabolite fingerprinting and metabolomic analysis of perchloric acid extracts from plant tissues. Nat. Protoc. 2008, 3, 1001–1012. [Google Scholar] [CrossRef]
- Lucarini, M.; Di Cocco, M.E.; Raguso, V.; Milanetti, F.; Durazzo, A.; Lombardi-Boccia, G.; Santini, A.; Delfini, M.; Sciubba, F. NMR-based metabolomic comparison of Brassica oleracea (var. italica): Organic and conventional farming. Foods 2020, 9, 945. [Google Scholar] [CrossRef]
- Hurtado, C.; Parastar, H.; Matamoros, V.; Piña, B.; Tauler, R.; Bayona, J.M. Linking the morphological and metabolomic response of Lactuca sativa L. exposed to emerging contaminants using GC× GC-MS and chemometric tools. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef]
- Kazimierczak, R.; Hallmann, E.; Lipowski, J.; Drela, N.; Kowalik, A.; Püssa, T.; Matt, D.; Luik, A.; Gozdowski, D.; Rembiałkowska, E. Beetroot (Beta vulgaris L.) and naturally fermented beetroot juices from organic and conventional production: Metabolomics, antioxidant levels and anticancer activity. J. Sci. Food Agric. 2014, 94, 2618–2629. [Google Scholar] [CrossRef] [PubMed]
- Hoult, D.I.; Busby, S.J.W.; Gadian, D.G.; Radda, G.K.; Richards, R.E.; Seeley, P.J. Observation of tissue metabolites using 31P nuclear magnetic resonance. Nature 1974, 252, 285–287. [Google Scholar] [CrossRef]
- Krishnan, P.; Kruger, N.J.; Ratcliffe, R.G. Metabolite fingerprinting and profiling in plants using NMR. J. Exp. Bot. 2005, 56, 255–265. [Google Scholar] [CrossRef] [Green Version]
- Crook, A.A.; Powers, R. Quantitative NMR-based biomedical metabolomics: Current status and applications. Molecules 2020, 25, 5128. [Google Scholar] [CrossRef] [PubMed]
- Moco, S.; Schneider, B.; Vervoort, J. Plant micro metabolomics: The analysis of endogenous metabolites present in a plant cell or tissue. J. Proteome Res. 2000, 8, 1694–1703. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Southam, A.D.; Hines, A.; Viant, M.R. High-throughput tissue extraction protocol for NMR-and MS-based metabolomics. Anal. Biochem. 2008, 372, 204–212. [Google Scholar] [CrossRef]
- Garcia-Perez, I.; Posma, J.M.; Serrano-Contreras, J.I.; Boulangé, C.L.; Chan, Q.; Frost, G.; Stamler, J.; Elliott, P.; Lindon, J.C.; Holmes, E.; et al. Identifying unknown metabolites using NMR-based metabolic profiling techniques. Nat. Protoc. 2020, 15, 2538–2567. [Google Scholar] [CrossRef]
- Fraga-Corral, M.; Carpena, M.; Garcia-Oliveira, P.; Pereira, A.G.; Prieto, M.A.; Simal-Gandara, J. Analytical metabolomics and applications in health, environmental and food science. Crit. Rev. Analytic. Chem. 2022, 52, 712–734. [Google Scholar] [CrossRef]
- Khakimov, B.; Bak, S.; Engelsen, S.B. High-throughput cereal metabolomics: Current analytical technologies, challenges and perspectives. J. Cereal Sci. 2014, 59, 393–418. [Google Scholar] [CrossRef]
- Bauermeister, A.; Mannochio-Russo, H.; Costa-Lotufo, L.V.; Jarmusch, A.K.; Dorrestein, P.C. Mass spectrometry-based metabolomics in microbiome investigations. Nat. Rev. Microbiol. 2022, 20, 143–160. [Google Scholar] [CrossRef]
- Sahoo, A.K.; Au, W.C.; Yang, C.S.; Mai, C.M.; Pan, C.L. THz Spectroscopy as Non-destructive Alternative to Secondary Ion Mass Spectroscopy. In Proceedings of the 45th International Conference on Infrared, Millimeter, and Terahertz Waves (IRMMW-THz), Buffalo, NY, USA, 8–13 November 2020; pp. 1–2. [Google Scholar] [CrossRef]
- Allen, F.; Pon, A.; Greiner, R.; Wishart, D. Computational prediction of electron ionization mass spectra to assist in GC/MS compound identification. Anal. Chem. 2016, 88, 7689–7697. [Google Scholar] [CrossRef]
- Vinaixa, M.; Schymanski, E.L.; Neumann, S.; Navarro, M.; Salek, R.M.; Yanes, O. Mass spectral databases for LC/MS-and GC/MS-based metabolomics: State of the field and future prospects. Trends Analytic. Chem. 2016, 78, 23–35. [Google Scholar] [CrossRef] [Green Version]
- Xiao, J.F.; Zhou, B.; Ressom, H.W. Metabolite identification and quantitation in LC-MS/MS-based metabolomics. Trends Anal. Chem. 2012, 32, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Majchrzak, T.; Wojnowski, W.; Lubinska-Szczygeł, M.; Różańska, A.; Namieśnik, J.; Dymerski, T. PTR-MS and GC-MS as complementary techniques for analysis of volatiles: A tutorial review. Anal. Chim. Acta 2018, 1035, 1–13. [Google Scholar] [CrossRef]
- Zhang, S.; Yin, F.; Li, J.; Ren, S.; Liang, X.; Zhang, Y.; Wang, L.; Wang, M.; Zhang, C. Transcriptomic and metabolomic investigation of metabolic disruption in Vigna unguiculata L. triggered by acetamiprid and cyromazine. Ecotoxicol. Environ. Safety 2022, 239, 113675. [Google Scholar] [CrossRef]
- Theodoridis, G.; Gika, H.G.; Wilson, I.D. LC-MS-based methodology for global metabolite profiling in metabonomics/metabolomics. Trends Analytic. Chem. 2008, 27, 251–260. [Google Scholar] [CrossRef]
- Cook, D.W.; Rutan, S.C.; Stoll, D.R.; Carr, P.W. Two-dimensional assisted liquid chromatography–a chemometric approach to improve accuracy and precision of quantitation in liquid chromatography using 2D separation, dual detectors, and multivariate curve resolution. Anal. Chim. Acta 2015, 859, 87–95. [Google Scholar] [CrossRef]
- Mondello, L.; Tranchida, P.Q.; Dugo, P.; Dugo, G. Comprehensive two-dimensional gas chromatography-mass spectrometry: A review. Mass Spectrom. Rev. 2008, 27, 101–124. [Google Scholar] [CrossRef]
- Mollerup, C.B.; Rasmussen, B.S.; Johansen, S.S.; Mardal, M.; Linnet, K.; Dalsgaard, P.W. Retrospective analysis for valproate screening targets with liquid chromatography–high resolution mass spectrometry with positive electrospray ionization: An omics-based approach. Drug Test. Anal. 2019, 11, 730–738. [Google Scholar] [CrossRef]
- Naz, S.; Gallart-Ayala, H.; Reinke, S.N.; Mathon, C.; Blankley, R.; Chaleckis, R.; Wheelock, C.E. Development of a liquid chromatography–high resolution mass spectrometry metabolomics method with high specificity for metabolite identification using all ion fragmentation acquisition. Anal. Chem. 2017, 89, 7933–7942. [Google Scholar] [CrossRef]
- Danek, M.; Plonka, J.; Barchanska, H. Metabolic profiles and non-targeted LC–MS/MS approach as a complementary tool to targeted analysis in assessment of plant exposure to pesticides. Food Chem. 2021, 356, 129680. [Google Scholar] [CrossRef] [PubMed]
- Anagnostopoulos, C.; Stasinopoulou, P.; Kanatas, P.; Travlos, I. Differences in metabolism of three Conyza species to herbicides glyphosate and triclopyr revealed by LC-MS/MS. Chil. J. Agric. Res. 2020, 80, 100–107. [Google Scholar] [CrossRef]
- González-Torralva, F.; Rojano-Delgado, A.M.; de Castro, M.D.L.; Mülleder, N.; De Prado, R. Two non-target mechanisms are involved in glyphosate-resistant horseweed (Conyza canadensis L. Cronq.) biotypes. J. Plant Physiol. 2012, 169, 1673–1679. [Google Scholar] [CrossRef]
- Yu, Y.; Wang, S.; Zhang, Q.; Yang, Y.; Chen, Y.; Liu, X.; Feng, C.; Hu, D.; Lu, P. Dissipation, residues, and risk assessment of imidacloprid in Zizania latifolia and purple sweet potato under field conditions using LC-MS/MS. J. Environ. Sci. Health Part B 2019, 54, 89–97. [Google Scholar] [CrossRef]
- Vu-Duc, N.; Nguyen-Quang, T.; Le-Minh, T.; Nguyen-Thi, X.; Tran, T.M.; Vu, H.A.; Nguyen, L.A.; Doan-Duy, T.; Van Hoi, B.; Vu, C.T.; et al. Multiresidue pesticides analysis of vegetables in Vietnam by ultrahigh-performance liquid chromatography in combination with high-resolution mass spectrometry (UPLC-Orbitrap MS). J. Anal. Methods Chem. 2019, 2019, 3489634. [Google Scholar] [CrossRef]
- Lee, H.; Depuydt, S.; Shin, K.; Choi, S.; Kim, G.; Lee, Y.H.; Park, J.T.; Han, T.; Park, J. Assessment of Various Toxicity Endpoints in Duckweed (Lemna minor) at the Physiological, Biochemical, and Molecular Levels as a Measure of Diuron Stress. Biology 2021, 10, 684. [Google Scholar] [CrossRef] [PubMed]
- Aliferis, K.A.; Materzok, S.; Paziotou, G.N.; Chrysayi-Tokousbalides, M. Lemna minor L. as a model organism for ecotoxicological studies performing 1H NMR fingerprinting. Chemosphere 2009, 76, 967–973. [Google Scholar] [CrossRef]
- Zhao, L.; Huang, Y.; Zhou, H.; Adeleye, A.S.; Wang, H.; Ortiz, C.; Mazer, S.J.; Keller, A.A. GC-TOF-MS based metabolomics and ICP-MS based metallomics of cucumber (Cucumis sativus) fruits reveal alteration of metabolites profile and biological pathway disruption induced by nano copper. Environ. Sci. Nano 2016, 3, 1114–1123. [Google Scholar] [CrossRef]
- Wu, L.; Gao, X.; Xia, F.; Joshi, J.; Borza, T.; Wang-Pruski, G. Biostimulant and fungicidal effects of phosphite assessed by GC-TOF-MS analysis of potato leaf metabolome. Physiol. Mol. Plant Pathol. 2019, 106, 49–56. [Google Scholar] [CrossRef]
- Zhao, L.; Hu, J.; Huang, Y.; Wang, H.; Adeleye, A.; Ortiz, C.; Keller, A.A. 1H NMR and GC–MS based metabolomics reveal nano-Cu altered cucumber (Cucumis sativus) fruit nutritional supply. Plant Physiol. Biochem. 2017, 110, 138–146. [Google Scholar] [CrossRef]
- Nagana Gowda, G.A.; Raftery, D. Recent advances in NMR-based metabolomics. Anal. Chem. 2017, 89, 490–510. [Google Scholar] [CrossRef]
- Zhang, A.; Sun, H.; Wang, P.; Han, Y.; Wang, X. Modern analytical techniques in metabolomics analysis. Analyst 2012, 137, 293–300. [Google Scholar] [CrossRef]
- Zhang, Y.; Huang, L.; Liu, L.; Cao, X.; Sun, C.; Lin, X. Metabolic disturbance in lettuce (Lactuca sativa) plants triggered by imidacloprid and fenvalerate. Sci. Total Environ. 2022, 802, 149764. [Google Scholar] [CrossRef] [PubMed]
- Gagnebin, Y.; Tonoli, D.; Lescuyer, P.; Ponte, B.; de Seigneux, S.; Martin, P.Y.; Schappler, J.; Boccard, J.; Rudaz, S. Metabolomic analysis of urine samples by UHPLC-QTOF-MS: Impact of normalization strategies. Anal. Chim. Acta 2017, 955, 27–35. [Google Scholar] [CrossRef]
- Fernandes, C.; Figueira, E.; Tauler, R.; Bedia, C. Exposure to chlorpyrifos induces morphometric, biochemical and lipidomic alterations in green beans (Phaseolus vulgaris). Ecotoxicol. Environ. Saf. 2018, 156, 25–33. [Google Scholar] [CrossRef]
- Ahsan, N.; Lee, D.G.; Lee, K.W.; Alam, I.; Lee, S.H.; Bahk, J.D.; Lee, B.H. Glyphosate-induced oxidative stress in rice leaves revealed by proteomic approach. Plant Physiol. Biochem. 2008, 46, 1062–1070. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Li, Q.; Zhao, J.; Peng, Y. Investigation of the effect of herbicide amiprophos methyl on spindle formation and proteome change in maize by immunofluorescence and proteomic technique. Cytologia 2011, 76, 249–259. [Google Scholar] [CrossRef]
- Fang, Y.; Lu, H.; Chen, S.; Zhu, K.; Song, H.; Qian, H. Leaf proteome analysis provides insights into the molecular mechanisms of bentazon detoxification in rice. Pestic. Biochem. Physiol. 2015, 125, 45–52. [Google Scholar] [CrossRef]
- Gholipour, Y.; Erra-Balsells, R.; Nonami, H. Detection of pesticides on tomato fruit surface by ultraviolet matrix-assisted laser desorption/ionization mass spectrometry. Environ. Control Biol. 2012, 50, 107–116. [Google Scholar] [CrossRef]
- Zhao, L.; Zhang, Y.; Wang, L.; Liu, X.; Zhang, J.; He, Z. Stereoselective metabolomic and lipidomic responses of lettuce (Lactuca sativa L.) exposing to chiral triazole fungicide tebuconazole. Food Chem. 2022, 371, 131209. [Google Scholar] [CrossRef]
- Lacina, O.; Urbanova, J.; Poustka, J.; Hajslova, J. Identification/quantification of multiple pesticide residues in food plants by ultra-high-performance liquid chromatography-time-of-flight mass spectrometry. J. Chromatogr. A 2010, 1217, 648–659. [Google Scholar] [CrossRef]
- Wang, J.; Wei, Q.; Wang, W.; Hu, H.; Yan, Y.; Wang, Y.; Li, Y.; Jiang, Y.; Wu, G.; Hu, T.; et al. Understanding the nutraceutical diversity through a comparative analysis of the taproot metabolomes of different edible radish types via UHPLC–Q–TOF–MS. Food Chem. 2023, 403, 134469. [Google Scholar] [CrossRef] [PubMed]
- Bauer, A.; Luetjohann, J.; Hanschen, F.S.; Schreiner, M.; Kuballa, J.; Jantzen, E.; Rohn, S. Identification and characterization of pesticide metabolites in Brassica species by liquid chromatography travelling wave ion mobility quadrupole time-of-flight mass spectrometry (UPLC-TWIMS-QTOF-MS). Food Chem. 2018, 244, 292–303. [Google Scholar] [CrossRef] [PubMed]
- Mu, Q.E.; Zhang, M.; Li, Y.; Feng, F.; Yu, X.; Nie, J. Metabolomic Analysis Reveals the Effect of Insecticide Chlorpyrifos on Rice Plant Metabolism. Metabolites 2022, 12, 1289. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Huang, Y.; Liu, J.; Chen, C.; Ouyang, G. In-vivo contaminant monitoring and metabolomic profiling in plants exposed to carbamates via a novel microextraction fiber. Environ. Sci. Tech. 2021, 55, 12449–12458. [Google Scholar] [CrossRef]
- Zhao, L.; Huang, Y.; Hu, J.; Zhou, H.; Adeleye, A.S.; Keller, A.A. 1H NMR and GC-MS based metabolomics reveal defense and detoxification mechanism of cucumber plant under nano-Cu stress. Environ. Sci. Tech. 2016, 50, 2000–2010. [Google Scholar] [CrossRef] [PubMed]
- Ge, G.; Jiao, W.; Cui, C.; Liao, G.; Sun, J.; Hou, R. Thiamethoxam metabolism and metabolic effects in cell suspension culture of tea (Camellia sinensis L.). J. Agric. Food Chem. 2019, 67, 7538–7546. [Google Scholar] [CrossRef] [PubMed]
- Boonchaisri, S.; Stevenson, T.; Dias, D.A. Utilization of GC–MS untargeted metabolomics to assess the delayed response of glufosinate treatment of transgenic herbicide resistant (HR) buffalo grasses (Stenotaphrum secundatum L.). Metabolomics 2020, 16, 1–25. [Google Scholar] [CrossRef]
- Ofoegbu, P.C.; Wagner, D.C.; Abolade, O.; Clubb, P.; Dobbs, Z.; Sayers, I.; Zenobio, J.E.; Adeleye, A.S.; Rico, C.M. Impacts of perfluorooctanesulfonic acid on plant biometrics and grain metabolomics of wheat (Triticum aestivum L.). J. Hazard. Mater. Adv. 2022, 7, 100131. [Google Scholar] [CrossRef]
- Emwas, A.H.M. The strengths and weaknesses of NMR spectroscopy and mass spectrometry with particular focus on metabolomics research. Metabonomics Methods Protoc. 2015, 1277, 161–193. [Google Scholar] [CrossRef]
- Scanlan, L.D.; Loguinov, A.V.; Teng, Q.; Antczak, P.; Dailey, K.P.; Nowinski, D.T.; Kornbluh, J.; Lin, X.X.; Lachenauer, E.; Arai, A.; et al. Gene transcription, metabolite and lipid profiling in eco-indicator Daphnia magna indicate diverse mechanisms of toxicity by legacy and emerging flame-retardants. Environ. Sci. Technol. 2015, 49, 7400–7410. [Google Scholar] [CrossRef] [PubMed] [Green Version]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shahid, M.; Singh, U.B.; Khan, M.S. Metabolomics-Based Mechanistic Insights into Revealing the Adverse Effects of Pesticides on Plants: An Interactive Review. Metabolites 2023, 13, 246. https://doi.org/10.3390/metabo13020246
Shahid M, Singh UB, Khan MS. Metabolomics-Based Mechanistic Insights into Revealing the Adverse Effects of Pesticides on Plants: An Interactive Review. Metabolites. 2023; 13(2):246. https://doi.org/10.3390/metabo13020246
Chicago/Turabian StyleShahid, Mohammad, Udai B. Singh, and Mohammad Saghir Khan. 2023. "Metabolomics-Based Mechanistic Insights into Revealing the Adverse Effects of Pesticides on Plants: An Interactive Review" Metabolites 13, no. 2: 246. https://doi.org/10.3390/metabo13020246
APA StyleShahid, M., Singh, U. B., & Khan, M. S. (2023). Metabolomics-Based Mechanistic Insights into Revealing the Adverse Effects of Pesticides on Plants: An Interactive Review. Metabolites, 13(2), 246. https://doi.org/10.3390/metabo13020246