Untargeted Metabolomics of Alternaria solani-Challenged Wild Tomato Species Solanum cheesmaniae Revealed Key Metabolite Biomarkers and Insight into Altered Metabolic Pathways
Abstract
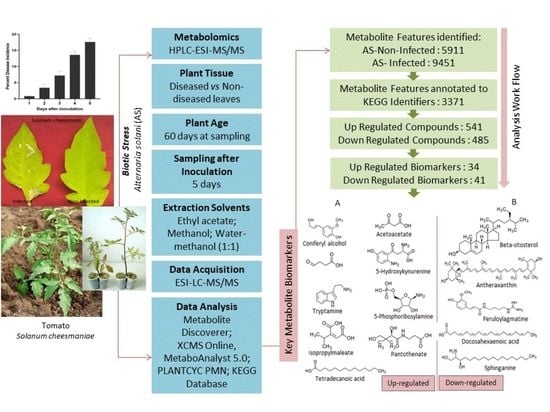
:1. Introduction
2. Materials and Methods
2.1. Plant Material and Growth Conditions
2.2. Leaf Sample Preparation and Metabolite Extraction
2.3. Untargeted LC–MS Analysis
2.4. Tandem MS Data Processing and Statistical Analysis
2.5. Statistical Analysis
3. Results
3.1. Enrichment Analysis
3.2. Univariate and Multivariate Data Analyses
3.3. Pathway Classification of Annotated Features by KEGG Mapper
3.4. Enrichment Analysis of Up- and Downregulated Compounds
3.5. Potential Upregulated Metabolites
3.6. Metabolite Biomarkers/Pathways for Resistance Response in S. cheesmaniae
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Iqbal, Z.; Iqbal, M.S.; Hashem, A.; Abd_Allah, E.F.; Ansari, M.I. Plant defense responses to biotic stress and its interplay with fluctuating dark/light conditions. Front. Plant Sci. 2021, 12, 631810. [Google Scholar] [CrossRef] [PubMed]
- Lamers, J.; Van Der Meer, T.; Testerink, C. How plants sense and respond to stressful environments. Plant Physiol. 2020, 182, 1624–1635. [Google Scholar] [CrossRef] [PubMed]
- Velásquez, A.C.; Castroverde, C.D.M.; He, S.Y. Plant-pathogen warfare under changing climate conditions. Curr. Biol. 2018, 28, R619–R634. [Google Scholar] [CrossRef] [PubMed]
- Saijo, Y.; Loo, E.P.I. Plant immunity in signal integration between biotic and abiotic stress responses. New Phytol. 2020, 225, 87–104. [Google Scholar] [CrossRef] [PubMed]
- Abdullah, A.S.; Moffat, C.S.; Lopez-Ruiz, F.J.; Gibberd, M.R.; Hamblin, J.; Zerihun, A. Host–multi-pathogen warfare: Pathogen interactions in co-infected plants. Front. Plant Sci. 2017, 8, 1806. [Google Scholar] [CrossRef] [PubMed]
- Schenk, P.M.; Carvalhais, L.C.; Kazan, K. Unraveling plant–microbe interactions: Can multi-species transcriptomics help? Trends Biotechnol. 2012, 30, 177–184. [Google Scholar] [CrossRef]
- Akamatsu, A.; Shimamoto, K.; Kawano, Y. Crosstalk of signalling mechanisms involved in host defense and symbiosis against microorganisms in rice. Curr. Genom. 2016, 17, 297–307. [Google Scholar] [CrossRef]
- Olive, A.J.; Sassetti, C.M. Metabolic crosstalk between host and pathogen: Sensing, adapting and competing. Nat. Rev. Microbiol. 2016, 14, 221–234. [Google Scholar] [CrossRef]
- Jan, R.; Asaf, S.; Numan, M.; Lubna; Kim, K.-M. Plant secondary metabolite biosynthesis and transcriptional regulation in response to biotic and abiotic stress conditions. Agronomy 2021, 11, 968. [Google Scholar] [CrossRef]
- Meena, K.K.; Sorty, A.M.; Bitla, U.M.; Choudhary, K.; Gupta, P.; Pareek, A.; Singh, D.P.; Prabha, R.; Sahu, P.K.; Gupta, V.K. Abiotic stress responses and microbe-mediated mitigation in plants: The Omics Strategies. Front. Plant Sci. 2017, 8, 172. [Google Scholar] [CrossRef]
- Seymour, G.B.; Rose, J.K.C. Tomato molecular biology—Special collection of papers for molecular horticulture. Mol. Hortic. 2022, 2, 21. [Google Scholar] [CrossRef]
- Lebeda, A.; Mieslerová, B.; Petřivalský, M.; Luhová, L.; Špundová, M.; Sedlářová, M.; Nožková-Hlaváčková, V.; Pink, D.A.C. Resistance mechanisms of wild tomato germplasm to infection of Oidiumneo lycopersici. Eur. J. Plant Pathol. 2014, 138, 569–596. [Google Scholar] [CrossRef]
- Pailles, Y.; Awlia, M.; Julkowska, M.; Passone, L.; Zemmouri, K.; Negrão, S.; Schmöckel, S.M.; Tester, M. Diverse traits contribute to salinity tolerance of wild tomato seedlings from the Galapagos Islands. Plant Physiol. 2020, 182, 534–546. [Google Scholar] [CrossRef] [PubMed]
- Yerasu, Y.R.; Murugan, L.; Halder, H.; Prasanna, H.C.; Singh, A.; Singh, B. Screening tomato genotypes for resistance to early blight and American Serpentine Leafminer. Hortic. Environ. Biotechnol. 2019, 60, 427–433. [Google Scholar] [CrossRef]
- Adhikari, P.; Oh, Y.; Panthee, D.R. Current status of early blight resistance in tomato: An update. Int. J. Mol. Sci. 2017, 18, 2019. [Google Scholar] [CrossRef] [PubMed]
- Allwood, J.W.; Williams, A.; Uthe, H.; van Dam, N.M.; Mur, L.A.J.; Grant, M.R.; Pétriacq, P. Unravelling plant responses to stress—The importance of targeted and untargeted metabolomics. Metabolites 2021, 11, 558. [Google Scholar] [CrossRef]
- Mushtaq, M.Y.; Choi, Y.H.; Verpoorte, R.; Wilson, E.G. Extraction for metabolomics: Access to the metabolome. Phytochem. Anal. 2014, 25, 291–306. [Google Scholar] [CrossRef]
- Jo, Y.-H.; Kim, S.; Kwon, S.-A.; Lee, H.J. Metabolomic analysis of ethyl acetate and methanol extracts of blueberry. J. Korean Soc. Food Sci. Nutr. 2014, 43, 419–424. [Google Scholar] [CrossRef]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.-M.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef]
- Sansone, S.A.; Fan, T.; Goodacre, R.; Griffin, J.L.; Hardy, N.W.; Kaddurah-Daouk, R.; Kristal, B.S.; Lindon, J.; Mendes, P.; Morrison, N.; et al. The metabolomics standards initiative. Nat. Biotechnol. 2007, 25, 846–848. [Google Scholar] [CrossRef]
- Rivera-Pérez, A.; Romero-González, R.; Garrido, F.A. Application of an innovative metabolomics approach to discriminate geographical origin and processing of black pepper by untargeted UHPLC-Q-Orbitrap-HRMS analysis and mid-level data fusion. Food Res. Int. 2021, 150A, 110722. [Google Scholar] [CrossRef] [PubMed]
- Quinet, M.; Angosto, T.; Yuste-Lisbona, F.J.; Blanchard-Gros, R.; Bigot, S.; Martinez, J.-P.; Lutts, S. Tomato fruit development and metabolism. Front. Plant Sci. 2019, 10, 1554. [Google Scholar] [CrossRef]
- Singh, D.P.; Bisen, M.S.; Shukla, R.; Prabha, R.; Maurya, S.; Reddy, Y.S.; Singh, P.M.; Rai, N.; Chaubey, T.; Chaturvedi, K.K.; et al. Metabolomics-driven mining of metabolite resources: Applications and prospects for improving vegetable crops. Int. J. Mol. Sci. 2022, 23, 12062. [Google Scholar] [CrossRef]
- Isah, T. Stress and defense responses in plant secondary metabolites production. Biol. Res. 2019, 52, 39. [Google Scholar] [CrossRef] [PubMed]
- Kachroo, A.; Kachroo, P. Fatty acid-derived signals in plant defense. Annu. Rev. Phytopathol. 2009, 47, 153–176. [Google Scholar] [CrossRef] [PubMed]
- Kishimoto, K.; Matsui, K.; Ozawa, R.; Takabayashi, J. Volatile C6-aldehydes and allo-ocimene activate defense genes and induce resistance against Botrytis cinerea in Arabidopsis thaliana. Plant Cell Physiol. 2005, 46, 1093–1102. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekaran, M.; Paramasivan, M.; Chun, S.C. Bacillus subtilis CBR05 induces vitamin B6 biosynthesis in tomato through the de novo pathway in contributing disease resistance against Xanthomonas campestris pv. Vesicatoria. Sci. Rep. 2019, 9, 6495. [Google Scholar] [CrossRef]
- Pushpalatha, H.B.; Mythrashree, S.R.; Shetty, R.; Geetha, N.; Sharathchandra, R.G.; Amruthesh, K.N.; Shetty, H.S. Ability of vitamins to induce downy mildew disease resistance and growth promotion in pearl millet. Crop Prot. 2007, 26, 1674–1681. [Google Scholar] [CrossRef]
- Ishihara, A.; Nakao, T.; Mashimo, Y.; Murai, M.; Ichimaru, N.; Tanaka, C.; Nakajima, H.; Wakasa, K.; Miyagawa, H. Probing the role of tryptophan-derived secondary metabolism in defense responses against Bipolaris oryzae infection in rice leaves by a suicide substrate of tryptophan decarboxylase. Phytochemistry 2011, 72, 7–13. [Google Scholar] [CrossRef]
- Hagemeier, J.; Schneider, B.; Oldham, N.J.; Hahlbrock, K. Accumulation of soluble and wall-bound indolic metabolites in Arabidopsis thaliana leaves infected with virulent or avirulent Pseudomonas syringae pathovar tomato strains. Proc. Natl. Acad. Sci. USA 2001, 98, 753–758. [Google Scholar] [CrossRef]
- Böttcher, C.; Chapman, A.; Fellermeier, F.; Choudhary, M.; Scheel, D.; Glawischnig, E. The biosynthetic pathway of indole-3-carboxaaldehyde and indole-3-carboxylic acid derivatives in Arabidopsis. Plant Physiol. 2014, 165, 841–853. [Google Scholar] [CrossRef] [PubMed]
- Shen, Q.; Liu, L.; Wang, L.; Wang, Q. Indole primes plant defense against necrotrophic fungal pathogen infection. PLoS ONE 2018, 13, e0207607. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, A.; Sood, P.; Citovsky, V. The roles of plant phenolics in defence and communication during Agrobacterium and Rhizobium infection. Mol. Plant Pathol. 2010, 11, 705–719. [Google Scholar] [CrossRef] [PubMed]
- Diaz-Tielase, C.; Grana, E.; Reigosa, M.J.; Sanchez-Moreiras, A.M. Biological activities and novel applications of chalcones. Plantadaninha 2016, 34, 607–616. [Google Scholar] [CrossRef]
- León, R.-H.; Hernández-Equihua, M.G.; Boone-Villa, D.; Jacobo, G.C.M.; Aguilera-Méndez, A. Biotin supplementation alters root system architecture and development in Arabidopsis thaliana. Plant Root 2019, 13, 29–40. [Google Scholar]
- Olorunnisola, O.S.; Ajayi, A.F.; Okeleji, L.O.; Oladipo, A.A.; Emorioloye, J.T. Vitamins as antioxidants. J. Food Sci. Nutr. Res. 2019, 2, 214–235. [Google Scholar]
- Völz, R.; Park, J.Y.; Harris, W.; Hwang, S.; Lee, Y.-H. Lyso-phosphatidylethanolamine primes the plant immune system and promotes basal resistance against hemibiotrophic pathogens. BMC Biotechnol. 2021, 21, 12. [Google Scholar] [CrossRef]
- Yu, M.H.; Zhao, Z.Z.; He, J.X. Brassinosteroid signaling in plant-microbe interactions. Int. J. Mol. Sci. 2018, 19, 4091. [Google Scholar] [CrossRef]
- Hout, B.; Yao, J.; Montgomery, B.L.; He, S.Y. Growth–Defense Tradeoffs in Plants: A Balancing Act to Optimize Fitness. Mol. Plant 2014, 7, 1267–1287. [Google Scholar] [CrossRef]
- Patocka, J.; Kuca, K. Biologically active alcohols: Cyclic alcohols. Mil. Med. Sci. Lett. Vojenske Zdr. Listy 2013, 82, 162–171. [Google Scholar] [CrossRef]
- Giner, R.M.; Ríos, J.L.; Máñez, S. Antioxidant activity of natural hydroquinones. Antioxidants 2022, 11, 343. [Google Scholar] [CrossRef]
- Wu, Y.; Liu, Y.; Gulbins, E.; Grassmé, H. The anti-infectious role of sphingosine in microbial diseases. Cells 2021, 10, 1105. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chen, Y.L.; Li, Y.K.; Chen, D.K.; He, J.F.; Yao, N. Functions of sphingolipids in pathogenesis during host–pathogen interactions. Front. Microbiol. 2021, 12, 701041. [Google Scholar] [CrossRef] [PubMed]
- Phung, T.H.; Jung, H.I.; Park, J.H.; Kim, J.G.; Back, K.; Jung, S. Porphyrin biosynthesis control under water stress: Sustained porphyrin status correlates with drought tolerance in transgenic rice. Plant Physiol. 2011, 157, 1746–1764. [Google Scholar] [CrossRef] [PubMed]
- Quesada, V.; Sarmiento-Mañús, R.; González-Bayón, R.; Hricová, A.; Ponce, M.R.; Micol, J.L. Porphobilinogen deaminase deficiency alters vegetative and reproductive development and causes lesions in Arabidopsis. PLoS ONE 2013, 8, e53378. [Google Scholar] [CrossRef] [PubMed]
- Aubry, S.; Fankhauser, N.; Ovinnikov, S.; Pružinská, A.; Stirnemann, M.; Zienkiewicz, K.; Herrfurth, C.; Feussner, I.; Hörtensteiner, S. Pheophorbide a may regulate jasmonate signaling during dark-induced senescence. Plant Physiol. 2020, 182, 776–791. [Google Scholar] [CrossRef]
- Hirashima, M.; Tanaka, R.; Tanaka, A. Light-independent cell death induced by accumulation of pheophorbide a in Arabidopsis thaliana. Plant Cell Physiol. 2009, 50, 719–729. [Google Scholar] [CrossRef]
- Chen, K.; Li, G.-J.; Bressan, R.A.; Song, C.-P.; Zhu, J.-K.; Zhao, Y. Abscisic acid dynamics, signaling, and functions in plants. J. Integr. Plant Biol. 2021, 62, 25–54. [Google Scholar] [CrossRef] [PubMed]
- Tal, L.; Gil, M.X.A.; Guercio, A.M.; Shabek, N. Structural aspects of plant hormone signal perception and regulation by ubiquitin ligases. Plant Physiol. 2020, 182, 1537–1544. [Google Scholar] [CrossRef]
- Jiménez-Bremont, J.F.; Marina, M.; de la Luz Guerrero-González, L.; Rossi, F.R.; Sánchez-Rangel, D.; Rodríguez-Kessler, M.; Ruiz, O.A.; Gárriz, A. Physiological and molecular implications of plant polyamine metabolism during biotic interactions. Front. Plant Sci. 2014, 18, 95. [Google Scholar] [CrossRef]
- Adio, A.M.; Casteel, C.L.; De Vos, M.; Kim, J.H.; Joshi, V.; Li, B.; Juéry, C.; Daron, J.; Kliebenstein, D.J.; Jander, G. Biosynthesis and defensive function of Nδ-acetylornithine, a jasmonate-induced Arabidopsis metabolite. Plant Cell 2011, 23, 3303–3318. [Google Scholar] [CrossRef]
- Zeier, J. New insights into the regulation of plant immunity by amino acid metabolic pathways. Plant Cell Environ. 2013, 36, 2085–2301. [Google Scholar] [CrossRef] [PubMed]
- Dye, S.M.; Yang, J.; Bostock, R.M. Eicosapolyenoic fatty acids alter oxylipin gene expression and fatty acid hydroperoxide profiles in tomato and pepper roots. Physiol. Mol. Plant Pathol. 2020, 109, 101444. [Google Scholar] [CrossRef]
- Yang, F.; Zhang, Q.; Yao, Q.; Chen, G.; Tong, H.; Zhang, J.; Su, Q.; Zhang, Y. Direct and indirect plant defenses induced by (Z)-3-hexenol in tomato against whitefly attack. J. Pest Sci. 2020, 93, 1243–1254. [Google Scholar] [CrossRef]
- Hatem, B.; Gargouri, M.; Mliki, A.; Brini, F.; Chong, J.; Moez, J. Vitamins for enhancing plant resistance. Planta 2016, 244, 529–543. [Google Scholar] [CrossRef]
- Swapnil, P.; Meena, M.; Singh, S.K.; Dhuldhaj, U.P.; Marwal, H.A. Vital roles of carotenoids in plants and humans to deteriorate stress with its structure, biosynthesis, metabolic engineering and functional aspects. Curr. Plant Biol. 2021, 26, 100203. [Google Scholar] [CrossRef]
- Sun, T.; Rao, S.; Zhou, X.; Li, L. Plant carotenoids: Recent advances and future perspectives. Mol. Hortic. 2022, 2, 3. [Google Scholar] [CrossRef]
- Rebelo, B.A.; Farrona, S.; Ventura, M.R.; Abranches, R. Canthaxanthin, a red-hot carotenoid: Applications, synthesis, and biosynthetic evolution. Plants 2020, 9, 1039. [Google Scholar] [CrossRef]
- Simkin, A.J. Carotenoids and apocarotenoids in planta: Their role in plant development, contribution to the flavour and aroma of fruits and flowers, and their nutraceutical benefits. Plants 2021, 10, 2321. [Google Scholar] [CrossRef]
- Latowski, D.; Kuczyńska, P.; Strzałka, K. Xanthophyll cycle—A mechanism protecting plants against oxidative stress. Redox Rep. 2011, 16, 78–90. [Google Scholar] [CrossRef]
- Liu, W.; Feng, Y.; Yu, S.; Fan, Z.; Li, X.; Li, J.; Yin, H. The flavonoid biosynthesis network in plants. Int. J. Mol. Sci. 2021, 22, 12824. [Google Scholar] [CrossRef] [PubMed]
- Geng, D.; Shen, X.; Xie, Y.; Yang, Y.; Bian, R.; Gao, Y.; Li, P.; Sun, L.; Feng, H.; Ma, F.; et al. Regulation of phenylpropanoid biosynthesis by MdMYB88 and MdMYB124 contributes to pathogen and drought resistance in apple. Hortic. Res. 2020, 7, 102. [Google Scholar] [CrossRef]
- Pun, M.; Khazanov, N.; Galsurker, O.; Weitman, M.; Kerem, Z.; Senderowitz, H.; Yedidia, I. Phloretin, an apple phytoalexin, affects the virulence and fitness of Pectobacterium brasiliense by interfering with quorum-sensing. Front. Plant Sci. 2021, 12, 671807. [Google Scholar] [CrossRef] [PubMed]
- Rogowska, A.; Szakiel, A. The role of sterols in plant response to abiotic stress. Phytochem. Rev. 2020, 19, 1525–1538. [Google Scholar] [CrossRef]
- Künstler, A.; Gullner, G.; Ádám, A.L.; Nagy, J.K.; Király, L. The versatile roles of sulfur-containing biomolecules in plant defense—A road to disease resistance. Plants 2020, 9, 1705. [Google Scholar] [CrossRef] [PubMed]
- Kaur, S.; Samota, M.K.; Choudhary, M.; Choudhary, M.; Pandey, A.K.; Sharma, A.; Thakur, J. How do plants defend themselves against pathogens-Biochemical mechanisms and genetic interventions. Physiol. Mol. Biol. Plant. 2022, 28, 485–504. [Google Scholar] [CrossRef] [PubMed]
- Castro-Moretti, F.R.; Gentzel, I.N.; Mackey, D.; Alonso, A.P. Metabolomics as an emerging tool for the study of plant–pathogen interactions. Metabolites 2020, 10, 52. [Google Scholar] [CrossRef] [PubMed]
- Pretorius, C.J.; Tugizimana, F.; Steenkamp, P.A.; Piater, L.A.; Dubery, I.A. Metabolomics for biomarker discovery: Key signatory metabolic profiles for the identification and discrimination of oat cultivars. Metabolites 2021, 11, 165. [Google Scholar] [CrossRef]










| S. No. | Mass (m/z) | Matched Compound | Putatively Identified Biomarker Metabolites * | Fold Change | VIP Score (OPLS DA) |
|---|---|---|---|---|---|
| 1. | 229.03 | C03090 | 5-Phosphoribosylamine | 11.99 | 1.22 |
| 2. | 278.12 | C00864 | Pantothenate | 8.98 | 1.02 |
| 3. | 203.06 | C02631 | 2-Isopropylmaleate | 7.51 | 1.34 |
| 4. | 203.06 | C00331 | Indolepyruvate | 7.50 | 1.34 |
| 5. | 161.04 | C00164 | Acetoacetate | 6.27 | 1.59 |
| 6. | 161.04 | C00232 | Succinate semialdehyde | 5.63 | 1.59 |
| 7. | 229.20 | C06424 | Tetradecanoic acid | 4.41 | 1.22 |
| 8. | 162.03 | C15606 | 1,2-Dihydroxy-5-(methylthio)pent-1-en-3-one | 4.32 | 1.58 |
| 9. | 228.08 | C00881 | Deoxycytidine | 4.21 | 1.23 |
| 10 | 220.10 | C00590 | Coniferyl alcohol | 3.8 | 1.26 |
| 11. | 243.09 | C00588 | Choline phosphate | 3.39 | 1.17 |
| 12. | 257.10 | C00670 | sn-Glycero-3-phosphocholine | 3.29 | 1.12 |
| 13. | 203.05 | C00331 | Indolepyruvate | 3.27 | 1.35 |
| 14. | 187.02 | C00121 | D-Ribose | 3.21 | 1.43 |
| 15. | 219.11 | C00398 | Tryptamine | 2.99 | 1.27 |
| 16. | 219.10 | C00437 | N-Acetylornithine | 2.99 | 1.27 |
| 17. | 247.02 | C00018 | Pyridoxal phosphate | 2.92 | 1.15 |
| 18. | 191.03 | C00438 | N-Carbamoyl-L-aspartate | 2.79 | 1.40 |
| 19. | 261.04 | C05817 | (1R,6R)-6-Hydroxy-2-succinylcyclohexa-2,4-diene-1-carboxylate | 2.65 | 1.09 |
| 20. | 145.09 | C08492 | 3-Hexenol | 2.62 | 1.19 |
| 21. | 271.09 | C00931 | Porphobilinogen | 2.59 | 1.06 |
| 22. | 169.05 | C13482 | Phosphodimethylethanolamine | 2.47 | 1.54 |
| 23. | 259.04 | C00352 | D-Glucosamine 6-phosphate | 2.36 | 1.11 |
| 24. | 257.06 | C09762 | Liquiritigenin | 2.32 | 1.12 |
| 25. | 277.12 | C00449 | N6-(L-1,3-Dicarboxypropyl)-L-lysine | 2.29 | 1.02 |
| 26. | 119.03 | C00033 | Acetate | 2.29 | 1.12 |
| 27. | 163.06 | C06001 | (S)-3-Hydroxyisobutyrate | 2.28 | 1.57 |
| 28. | 146.04 | C00423 | trans-Cinnamate | 2.24 | 1.16 |
| 29. | 168.05 | C00436 | N-Carbamoylputrescine | 2.24 | 1.55 |
| 30. | 229.03 | C03090 | 5-Phosphoribosylamine | 2.18 | 1.23 |
| 31. | 261.11 | C00140 | N-Acetyl-D-glucosamine | 2.15 | 1.09 |
| 32. | 247.13 | C13455 | Abscisic aldehyde | 2.14 | 1.14 |
| 33. | 259.08 | C00120 | Biotin | 2.14 | 1.11 |
| 34. | 231.05 | C16361 | 1,3,7-Trimethyluric acid | 2.13 | 1.22 |
| S. No. | Mass (m/z) | Matched Compound | Putatively Identified Biomarker Metabolites * | Fold Change | VIP Score (OPLS DA) |
|---|---|---|---|---|---|
| 1. | 429.37 | C01753 | beta-Sitosterol | 0.403 | 1.89 |
| 2. | 419.08 | C00448 | trans,trans-Farnesyldiphosphate | 0.079 | 1.84 |
| 3. | 413.34 | C15777 | Episterol | 0.297 | 1.87 |
| 4. | 399.33 | C05437 | Zymosterol | 0.001 | 1.76 |
| 5. | 609.27 | C18098 | Primary fluorescent chlorophyll catabolite | 0.002 | 1.75 |
| 6. | 565.41 | C08579 | Antheraxanthin | 0.002 | 1.75 |
| 7. | 318.21 | C00909 | Leukotriene A4 | 0.009 | 1.75 |
| 8. | 337.24 | C06427 | (9Z,12Z,15Z)-Octadecatrienoic acid | 0.019 | 1.75 |
| 9. | 357.26 | C19616 | 18-Hydroxyoleate | 0.002 | 1.73 |
| 10 | 565.39 | C08583 | Canthaxanthin | 0.003 | 1.67 |
| 11. | 293.21 | C18218 | 16-Hydroxypalmitate | 0.005 | 1.67 |
| 12. | 417.34 | C15882 | 2-Methyl-6-phytylquinol | 0.007 | 1.66 |
| 13. | 489.39 | C02477 | alpha-Tocopherol | 0.004 | 1.65 |
| 14. | 417.34 | C15882 | 2-Methyl-6-phytylquinol | 0.008 | 1.63 |
| 15. | 434.27 | C05797 | Pheophytin a | 0.009 | 1.59 |
| 16. | 439.36 | C22121 | Cycloeucalenone | 0.011 | 1.58 |
| 17. | 329.27 | C01530 | Octadecanoic acid | 0.007 | 1.57 |
| 18. | 311.22 | C04717 | (9Z,11E)-(13S)-13-Hydroperoxyoctadeca-9,11-dienoic acid | 0.43 | 1.57 |
| 19. | 339.16 | C20693 | Carlactone | 0.321 | 1.56 |
| 20. | 380.35 | C08281 | Docosanoic acid | 0.172 | 1.54 |
| 21. | 549.41 | C06098 | Zeaxanthin | 0.011 | 1.54 |
| 22. | 423.36 | C01943 | Obtusifoliol | 0.011 | 1.47 |
| 23. | 337.04 | C00364 | dTMP | 0.441 | 1.46 |
| 24. | 324.10 | C01762 | Xanthosine | 0.374 | 1.44 |
| 25. | 180.10 | C05332 | Phenethylamine | 0.032 | 1.44 |
| 26. | 491.21 | C05427 | Phytyldiphosphate | 0.012 | 1.42 |
| 27. | 411.38 | C00751 | Squalene | 0.213 | 1.37 |
| 28. | 344.28 | C02934 | 3-Dehydrosphinganine | 0.131 | 1.36 |
| 29. | 282.28 | C00836 | Sphinganine | 0.432 | 1.26 |
| 30. | 198.08 | C02631 | 2-Isopropylmaleate | 0.441 | 1.23 |
| 31. | 337.20 | C01226 | 12-OPDA | 0.431 | 1.22 |
| 32. | 324.20 | C19691 | Farnesylcysteine | 0.206 | 1.21 |
| 33. | 309.19 | C05819 | Menaquinol | 0.314 | 1.18 |
| 34. | 383.10 | C20693 | Carlactone | 0.031 | 1.17 |
| 35. | 344.28 | C02934 | 3-Dehydrosphinganine | 0.043 | 1.12 |
| 36. | 427.14 | C01674 | N,N-diacetylchitobiose | 0.434 | 1.11 |
| 37. | 217.05 | C00794 | D-Sorbitol | 0.251 | 1.11 |
| 38. | 453.33 | C15800 | 3-Dehydro-6-deoxoteasterone | 0.011 | 1.09 |
| 39. | 493.28 | C18217 | 16-Feruloyloxypalmitate | 0.031 | 1.09 |
| 40. | 423.36 | C01943 | Obtusifoliol | 0.032 | 1.06 |
| 41. | 441.07 | C10646 | Lariciresinol | 0.491 | 1.02 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Singh, D.P.; Bisen, M.S.; Prabha, R.; Maurya, S.; Yerasu, S.R.; Shukla, R.; Tiwari, J.K.; Chaturvedi, K.K.; Farooqi, M.S.; Srivastava, S.; et al. Untargeted Metabolomics of Alternaria solani-Challenged Wild Tomato Species Solanum cheesmaniae Revealed Key Metabolite Biomarkers and Insight into Altered Metabolic Pathways. Metabolites 2023, 13, 585. https://doi.org/10.3390/metabo13050585
Singh DP, Bisen MS, Prabha R, Maurya S, Yerasu SR, Shukla R, Tiwari JK, Chaturvedi KK, Farooqi MS, Srivastava S, et al. Untargeted Metabolomics of Alternaria solani-Challenged Wild Tomato Species Solanum cheesmaniae Revealed Key Metabolite Biomarkers and Insight into Altered Metabolic Pathways. Metabolites. 2023; 13(5):585. https://doi.org/10.3390/metabo13050585
Chicago/Turabian StyleSingh, Dhananjaya Pratap, Mansi Singh Bisen, Ratna Prabha, Sudarshan Maurya, Suresh Reddy Yerasu, Renu Shukla, Jagesh Kumar Tiwari, Krishna Kumar Chaturvedi, Md. Samir Farooqi, Sudhir Srivastava, and et al. 2023. "Untargeted Metabolomics of Alternaria solani-Challenged Wild Tomato Species Solanum cheesmaniae Revealed Key Metabolite Biomarkers and Insight into Altered Metabolic Pathways" Metabolites 13, no. 5: 585. https://doi.org/10.3390/metabo13050585
APA StyleSingh, D. P., Bisen, M. S., Prabha, R., Maurya, S., Yerasu, S. R., Shukla, R., Tiwari, J. K., Chaturvedi, K. K., Farooqi, M. S., Srivastava, S., Rai, A., Sarma, B. K., Rai, N., Singh, P. M., Behera, T. K., & Farag, M. A. (2023). Untargeted Metabolomics of Alternaria solani-Challenged Wild Tomato Species Solanum cheesmaniae Revealed Key Metabolite Biomarkers and Insight into Altered Metabolic Pathways. Metabolites, 13(5), 585. https://doi.org/10.3390/metabo13050585