NblA1/A2-Dependent Homeostasis of Amino Acid Pools during Nitrogen Starvation in Synechocystis sp. PCC 6803
Abstract
:1. Introduction
2. Results and Discussion
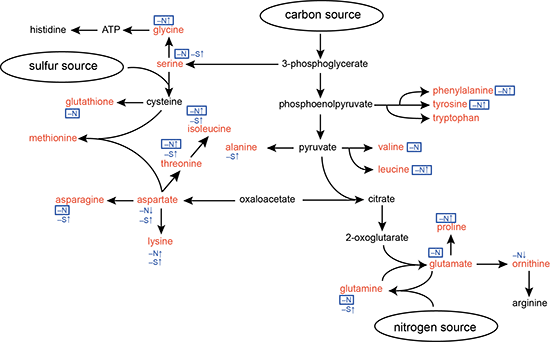
2.1. Responses of Amino Acid Pools to Nitrogen Starvation in Synechocystis Cells


2.2. Responses to Nitrogen Repletion following Starvation

2.3. Response of the nblA1/A2 Mutant to Nitrogen Starvation



2.4. Response of the nblA1/A2 Mutant to Sulfur Starvation

2.5. NblA1/A2–Dependent and –Independent Homeostasis of Amino Acids
3. Experimental Section
3.1. Culture Conditions and Cell Sampling
3.2. Disruption of nblA1/nblA2
3.3. Extraction of Amino Acids
3.4. Derivatization and GC-MS Analysis of Amino Acids
3.5. Bradford Assay
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Schwarz, R.; Forchhammer, K. Acclimation of unicellular cyanobacteria to macronutrient deficiency: Emergence of a complex network of cellular responses. Microbiology 2005, 151, 2503–2514. [Google Scholar] [CrossRef]
- Grossman, A. Acclimation of Chlamydomonas reinhardtii to its nutrient environment. Protist 2000, 151, 201–224. [Google Scholar] [CrossRef]
- Markou, G.; Nerantzis, E. Microalgae for high-value compounds and biofuels production: A review with focus on cultivation under stress conditions. Biotechnol. Adv. 2013, 31, 1532–1542. [Google Scholar] [CrossRef]
- Toepel, J.; Albaum, S.P.; Arvidsson, S.; Grossman, A.; la Russa, M.; Rogge, K.; Kruse, O. Construction and evaluation of a whole genome microarray of Chlamydomonas reinhardtii. BMC Genomics 2011, 12, 579. [Google Scholar] [CrossRef]
- Mitschke, J.; Vioque, A.; Haas, F.; Hess, W.R.; Muro-Pastor, A.M. Dynamics of transcriptional start site selection during nitrogen stress-induced cell differentiation in Anabaena sp. PCC7120. Proc. Natl. Acad. Sci. USA 2011, 108, 20130–20135. [Google Scholar]
- Dagenais-Bellefeuille, S.; Morse, D. Putting the N in dinoflagellates. Front. Microbiol. 2013, 4, 369. [Google Scholar]
- Ito, T.; Tanaka, M.; Shinkawa, H.; Nakada, T.; Ano, Y.; Kurano, N.; Soga, T.; Tomita, M. Metabolic and morphological changes of an oil accumulating trebouxiophycean alga in nitrogen-deficient conditions. Metabolomics 2013, 9, 178–187. [Google Scholar] [CrossRef]
- Dong, H.P.; Williams, E.; Wang, D.Z.; Xie, Z.X.; Hsia, R.C.; Jenck, A.; Halden, R.; Li, J.; Chen, F.; Place, A.R. Responses of Nannochloropsis oceanica IMET1 to long-term nitrogen starvation and recovery. Plant Physiol. 2013, 162, 1110–1126. [Google Scholar] [CrossRef]
- Watanabe, M.; Ikeuchi, M. Phycobilisome: Architecture of a light-harvesting supercomplex. Photosynth. Res. 2013, 116, 265–276. [Google Scholar] [CrossRef]
- Collier, J.L.; Grossman, A.R. Chlorosis induced by nutrient deprivation in Synechococcus sp. strain PCC 7942: Not all bleaching is the same. J. Bacteriol. 1992, 174, 4718–4726. [Google Scholar]
- Collier, J.L.; Grossman, A.R. A small polypeptide triggers complete degradation of light-harvesting phycobiliproteins in nutrient-deprived cyanobacteria. EMBO J. 1994, 13, 1039–1047. [Google Scholar]
- Li, H.; Sherman, L.A. Characterization of Synechocystis sp. strain PCC 6803 and deltanbl mutants under nitrogen-deficient conditions. Arch. Microbiol. 2002, 178, 256–266. [Google Scholar] [CrossRef]
- Karradt, A.; Sobanski, J.; Mattow, J.; Lockau, W.; Baier, K. NblA, a key protein of phycobilisome degradation, interacts with ClpC, a HSP100 chaperone partner of a cyanobacterial Clp protease. J. Biol. Chem. 2008, 283, 32394–32403. [Google Scholar]
- Hasunuma, T.; Kikuyama, F.; Matsuda, M.; Aikawa, S.; Izumi, Y.; Kondo, A. Dynamic metabolic profiling of cyanobacterial glycogen biosynthesis under conditions of nitrate depletion. J. Exp. Bot. 2013, 64, 2943–2954. [Google Scholar] [CrossRef]
- Coronil, T.; Lara, C. Amino acid level and expression of the nitrate assimilation system in Anacystis nidulans cells. Plant Physiol. Biochem. 1991, 29, 651–655. [Google Scholar]
- Osanai, T.; Oikawa, A.; Shirai, T.; Kuwahara, A.; Iijima, H.; Tanaka, K.; Ikeuchi, M.; Kondo, A.; Saito, K.; Hirai, M.Y. Capillary electrophoresis-mass spectrometry reveals the distribution of carbon metabolites during nitrogen starvation in Synechocystis sp. PCC 6803. Environ. Microbiol. 2014, 16, 512–524. [Google Scholar] [CrossRef]
- Hauf, W.; Schlebusch, M.; Hüge, J.; Kopka, J.; Hagemann, M.; Forchhammer, K. Metabolic changes in Synechocystis PCC6803 upon nitrogen-starvation: excess NADPH sustains polyhydroxybutyrate accumulation. Metabolites 2013, 3, 101–118. [Google Scholar] [CrossRef]
- Kaneko, T.; Sato, S.; Kotani, H.; Tanaka, A.; Asamizu, E.; Nakamura, Y.; Miyajima, N.; Hirosawa, M.; Sugiura, M.; Sasamoto, S.; et al. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions (supplement). DNA Res. 1996, 3, 185–209. [Google Scholar] [CrossRef]
- Yoshikawa, K.; Hirasawa, T.; Ogawa, K.; Hidaka, Y.; Nakajima, T.; Furusawa, C.; Shimizu, H. Integrated transcriptomic and metabolomic analysis of the central metabolism of Synechocystis sp. PCC 6803 under different trophic conditions. Biotechnol. J. 2013, 8, 571–580. [Google Scholar] [CrossRef]
- Kanesaki, Y.; Shiwa, Y.; Tajima, N.; Suzuki, M.; Watanabe, S.; Sato, N.; Ikeuchi, M.; Yoshikawa, H. Identification of substrain-specific mutations by massively parallel whole-genome resequencing of Synechocystis sp. PCC 6803. DNA Res. 2012, 19, 67–79. [Google Scholar] [CrossRef]
- Eisenhut, M.; Huege, J.; Schwarz, D.; Bauwe, H.; Kopka, J.; Hagemann, M. Metabolome phenotyping of inorganic carbon limitation in cells of the wild type and photorespiratory mutants of the cyanobacterium Synechocystis sp. strain PCC 6803. Plant Physiol. 2008, 148, 2109–2120. [Google Scholar] [CrossRef]
- Takahashi, H.; Uchimiya, H.; Hihara, Y. Difference in metabolite levels between photoautotrophic and photomixotrophic cultures of Synechocystis sp. PCC 6803 examined by capillary electrophoresis electrospray ionization mass spectrometry. J. Exp. Bot. 2008, 59, 3009–3018. [Google Scholar] [CrossRef]
- Liu, X.; Sheng, J.; Curtiss, R. Fatty acid production in genetically modified cyanobacteria. Proc. Natl. Acad. Sci. USA 2011, 108, 6899–6904. [Google Scholar] [CrossRef]
- Bentley, F.K.; Melis, A. Diffusion-based process for carbon dioxide uptake and isoprene emission in gaseous/aqueous two-phase photobioreactors by photosynthetic microorganisms. Biotechnol. Bioeng. 2012, 109, 100–109. [Google Scholar] [CrossRef]
- Atsumi, S.; Higashide, W.; Liao, J.C. Direct photosynthetic recycling of carbon dioxide to isobutyraldehyde. Nat. Biotechnol. 2009, 27, 1177–1180. [Google Scholar] [CrossRef]
- Kuniyoshi, T.M.; Gonzalez, A.; Lopez-Gomollon, S.; Valladares, A.; Bes, M.T.; Fillat, M.F.; Peleato, M.L. 2-oxoglutarate enhances NtcA binding activity to promoter regions of the microcystin synthesis gene cluster. FEBS Lett. 2011, 585, 3921–3926. [Google Scholar]
- Ohashi, Y.; Shi, W.; Takatani, N.; Aichi, M.; Maeda, S.; Watanabe, S.; Yoshikawa, H.; Omata, T. Regulation of nitrate assimilation in cyanobacteria. J. Exp. Bot. 2011, 62, 1411–1424. [Google Scholar] [CrossRef]
- Baier, K.; Nicklisch, S.; Grundner, C.; Reinecke, J.; Lockau, W. Expression of two nblA-homologous genes is required for phycobilisome degradation in nitrogen-starved Synechocystis sp. PCC 6803. FEMS Microbiol. Lett. 2001, 195, 35–39. [Google Scholar] [CrossRef]
- Grossman, A.R.; Schaefer, M.R.; Chiang, G.G.; Collier, J.L. The phycobilisome, a light-harvesting complex responsive to environmental conditions. Microbiol. Rev. 1993, 57, 725–749. [Google Scholar]
- Kiyota, H.; Ikeuchi, M.; Hirai, M.Y. Response of amino acid metabolism to sulfur starvation in Synechocystis sp. PCC 6803. In Sulfur Metabolism in Plants; Springer: Dordrecht, Netherlands, 2012; pp. 53–59. [Google Scholar]
- Bennett, B.D.; Kimball, E.H.; Gao, M.; Osterhout, R.; Van Dien, S.J.; Rabinowitz, J.D. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat. Chem. Biol. 2009, 5, 593–599. [Google Scholar] [CrossRef]
- Hans, M.A.; Heinzle, E.; Wittmann, C. Quantification of intracellular amino acids in batch cultures of Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2001, 56, 776–779. [Google Scholar] [CrossRef]
- Field, B.; Cardon, G.; Traka, M.; Botterman, J.; Vancanneyt, G.; Mithen, R. Glucosinolate and amino acid biosynthesis in Arabidopsis. Plant Physiol. 2004, 135, 828–839. [Google Scholar]
- Richaud, C.; Zabulon, G.; Joder, A.; Thomas, J.C. Nitrogen or sulfur starvation differentially affects phycobilisome degradation and expression of the nblA gene in Synechocystis strain PCC 6803. J. Bacteriol. 2001, 183, 2989–2994. [Google Scholar] [CrossRef]
- KEGG Lysine degradation—Synechocystis sp. PCC 6803. Available online: http://www.genome.jp/kegg-bin/show_pathway?syn00310 (accessed on 26 June 2014).
- Rippka, R. Isolation and purification of cyanobacteria. Methods Enzymol. 1988, 167, 3–27. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Kiyota, H.; Hirai, M.Y.; Ikeuchi, M. NblA1/A2-Dependent Homeostasis of Amino Acid Pools during Nitrogen Starvation in Synechocystis sp. PCC 6803. Metabolites 2014, 4, 517-531. https://doi.org/10.3390/metabo4030517
Kiyota H, Hirai MY, Ikeuchi M. NblA1/A2-Dependent Homeostasis of Amino Acid Pools during Nitrogen Starvation in Synechocystis sp. PCC 6803. Metabolites. 2014; 4(3):517-531. https://doi.org/10.3390/metabo4030517
Chicago/Turabian StyleKiyota, Hiroshi, Masami Yokota Hirai, and Masahiko Ikeuchi. 2014. "NblA1/A2-Dependent Homeostasis of Amino Acid Pools during Nitrogen Starvation in Synechocystis sp. PCC 6803" Metabolites 4, no. 3: 517-531. https://doi.org/10.3390/metabo4030517
APA StyleKiyota, H., Hirai, M. Y., & Ikeuchi, M. (2014). NblA1/A2-Dependent Homeostasis of Amino Acid Pools during Nitrogen Starvation in Synechocystis sp. PCC 6803. Metabolites, 4(3), 517-531. https://doi.org/10.3390/metabo4030517