Metabolomic Markers for the Early Selection of Coffea canephora Plants with Desirable Cup Quality Traits
Abstract
:1. Introduction
2. Results and Discussion
2.1. Variability of Metabolic Phenotypes and the Relationship Between C. Canephora Parent Plants
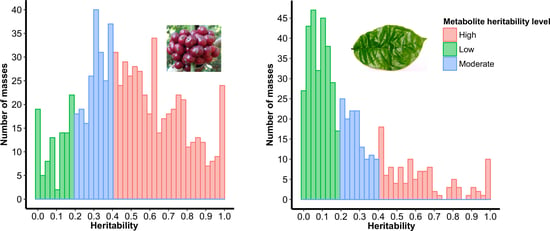
2.2. Selection of Highly Heritable Metabolites in Coffee Fruits and Leaves of C. Canephora Families
2.3. Metabolites of Roasted Coffee Beans Linked to Sensory Traits of Coffee Beverage
2.3.1. Tasting and Correlation Between Sensory Attributes of Coffee Beverage
2.3.2. Selection of Marker Metabolites in Roasted Coffee Beans Linked to Sensory Traits
2.4. Highly Heritable Metabolites in Fruits Linked to Coffee Cup Quality
2.5. Highly Heritable Metabolites in Leaf Linked to Coffee Cup Quality
3. Materials and Methods
3.1. Heritability Analysis
3.1.1. Leaves and Fruits Sampling
3.1.2. Leaves and Fruits Tissue Metabolite Extraction
3.1.3. Estimation of Metabolites Heritability
3.2. UPLC-ESI-MS Analysis
3.3. Spectra Data Analysis and Putative Identification of Metabolites
3.4. Cup Quality-Related Metabolite Analysis
3.4.1. Coffee Beans Processing and Sensory Analysis of Coffee Beverage
3.4.2. Roasted Beans Metabolite Extraction
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Esquivel, P.; Jiménez, V.M. Functional properties of coffee and coffee by-products. Food Res. Int. 2012, 46, 488–495. [Google Scholar] [CrossRef]
- Farah, A. 15 - Coffee as a speciality and functional beverage A2 - Paquin, Paul. In Functional and Speciality Beverage Technology; Woodhead Publishing Series in Food Science, Technology and Nutrition; Woodhead Publishing: Cambridge, UK, 2009; pp. 370–395. ISBN 978-1-84569-342-8. [Google Scholar]
- Charrier, A.; Eskes, A.B. Botany and Genetics of Coffee. In Coffee: Growing, Processing, Sustainable Production; Wiley-VCH: Weinheim, Germany, 2008; pp. 25–56. ISBN 978-3-527-30731-9. [Google Scholar]
- Leroy, T.; Ribeyre, F.; Bertrand, B.; Charmetant, P.; Dufour, M.; Montagnon, C.; Marraccini, P.; Pot, D. Genetics of coffee quality. Braz. J. Plant Physiol. 2006, 18, 229–242. [Google Scholar] [CrossRef]
- Montagnon, C.; Marraccini, P.; Bertrand, B. Breeding for coffee quality. In Specialty Coffee: Managing Quality, Oberthür, T., Läderach, P., Pohlan, H.A.J., Cock, J., Eds.; IPNI: Southeast Asia, 2012; pp. 89–117. [Google Scholar]
- Philippe, L.; Benoít, B.; Hervé, E. Breeding coffee (Coffea arabica) for sustainable production. In Breeding Plantation Tree Crops: Tropical Species; Springer: Berlin/Heidelberg, Germany, 2009; pp. 525–543. [Google Scholar]
- Wintgens, J.N. Coffee: Growing, Processing, Sustainable Production. Available online: http://onlinelibrary.wiley.com/book/10.1002/9783527619627 (accessed on 23 March 2016).
- Leroy, T.; De Bellis, F.; Legnate, H.; Kananura, E.; Gonzales, G.; Pereira, L.F.; Andrade, A.C.; Charmetant, P.; Montagnon, C.; Cubry, P.; et al. Improving the quality of African robustas: QTLs for yield- and quality-related traits in Coffea canephora. Tree Genet. Genomes 2011, 7, 781–798. [Google Scholar] [CrossRef]
- Davis, A.P.; Gole, T.W.; Baena, S.; Moat, J. The Impact of Climate Change on Indigenous Arabica Coffee (Coffea arabica): Predicting Future Trends and Identifying Priorities. PLoS ONE 2012, 7, e47981. [Google Scholar] [CrossRef] [PubMed]
- Silva, M.D.C.; Várzea, V.; Guerra-Guimarães, L.; Azinheira, H.G.; Fernandez, D.; Petitot, A.-S.; Bertrand, B.; Lashermes, P.; Nicole, M. Coffee resistance to the main diseases: leaf rust and coffee berry disease. Braz. J. Plant Physiol. 2006, 18, 119–147. [Google Scholar] [CrossRef] [Green Version]
- Santos, C.; Leitao, A.; Pais, I.; Lidon, F.; Ramalho, J. Perspectives on the potential impacts of climate changes on coffee plant and bean quality. Emir. J. Food Agric. 2015, 27, 152. [Google Scholar] [CrossRef]
- Corso, M.P.; Vignoli, J.A.; Benassi, M. de T. Development of an instant coffee enriched with chlorogenic acids. J. Food Sci. Technol. 2016, 53, 1380–1388. [Google Scholar] [CrossRef]
- Farah, A. Coffee Constituents. In Coffee; Chu, Y.-F., Ed.; Wiley-Blackwell: Hoboken, NJ, USA, 2012; pp. 21–58. ISBN 978-1-119-94989-3. [Google Scholar]
- Akaffou, D.S.; Hamon, P.; Doulbeau, S.; Keli, J.; Legnate, H.; Campa, C.; Hamon, S.; de Kochko, A.; Zoro, B.I.A. Inheritance and relationship between key agronomic and quality traits in an interspecific cross between Coffea pseudozanguebariae Bridson and C. canephora Pierre. Tree Genet. Genomes 2012, 8, 1149–1162. [Google Scholar] [CrossRef]
- Ky, C.-L.; Louarn, J.; Guyot, B.; Charrier, A.; Hamon, S.; Noirot, M. Relations between and inheritance of chlorogenic acid contents in an interspecific cross between Coffea pseudozanguebariae and Coffea liberica var “dewevrei”. Theor. Appl. Genet. 1999, 98, 628–637. [Google Scholar] [CrossRef]
- Ky, C.-L.; Guyot, B.; Louarn, J.; Hamon, S.; Noirot, M. Trigonelline inheritance in the interspecific Coffea pseudozanguebariae×C. liberica var. dewevrei cross. Theor. Appl. Genet. 2001, 102, 630–634. [Google Scholar] [CrossRef]
- Mérot-L’Anthoëne, V.; Mangin, B.; Lefebvre-Pautigny, F.; Jasson, S.; Rigoreau, M.; Husson, J.; Lambot, C.; Crouzillat, D. Comparison of three QTL detection models on biochemical, sensory, and yield characters in Coffea canephora. Tree Genet. Genomes 2014, 10, 1541–1553. [Google Scholar] [CrossRef]
- Montagnon, C.; Guyot, B.; Cilas, C.; Leroy, T. Genetic parameters of several biochemical compounds from green coffee, Coffea canephora. Plant Breed. 1998, 117, 576–578. [Google Scholar] [CrossRef]
- Tran, H.T.; Lee, L.S.; Furtado, A.; Smyth, H.; Henry, R.J. Advances in genomics for the improvement of quality in coffee: Genomics for improving coffee quality. J. Sci. Food Agric. 2016, 96, 3300–3312. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, O.; Urrutia, M.; Bernillon, S.; Giauffret, C.; Tardieu, F.; Le Gouis, J.; Langlade, N.; Charcosset, A.; Moing, A.; Gibon, Y. Fortune telling: metabolic markers of plant performance. Metabolomics 2016, 12. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Bohra, A.; Pandey, A.K.; Pandey, M.K.; Kumar, A. Metabolomics for Plant Improvement: Status and Prospects. Front. Plant Sci. 2017, 8, 1302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cuperlovic-Culf, M.; Wang, L.; Forseille, L.; Boyle, K.; Merkley, N.; Burton, I.; Fobert, P.R. Metabolic Biomarker Panels of Response to Fusarium Head Blight Infection in Different Wheat Varieties. PloS ONE 2016, 11, e0153642. [Google Scholar] [CrossRef] [PubMed]
- Heuberger, A.L.; Broeckling, C.D.; Kirkpatrick, K.R.; Prenni, J.E. Application of nontargeted metabolite profiling to discover novel markers of quality traits in an advanced population of malting barley. Plant Biotechnol. J. 2014, 12, 147–160. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.W.; Kim, H.-T.; Lee, W.Y.; Choi, M.N.; Park, E.-J. Identification of a potential metabolic marker, inositol, for the inherently fast growth trait by stems of Pinus densiflora via a retrospective approach. Can. J. For. Res. 2015, 45, 770–775. [Google Scholar] [CrossRef]
- Lima, M.R.M.; Felgueiras, M.L.; Graça, G.; Rodrigues, J.E.A.; Barros, A.; Gil, A.M.; Dias, A.C.P. NMR metabolomics of esca disease-affected Vitis vinifera cv. Alvarinho leaves. J. Exp. Bot. 2010, 61, 4033–4042. [Google Scholar] [CrossRef]
- López, M.G.; Zanor, M.I.; Pratta, G.R.; Stegmayer, G.; Boggio, S.B.; Conte, M.; Bermúdez, L.; Coluccio Leskow, C.; Rodríguez, G.R.; Picardi, L.A.; et al. Metabolic analyses of interspecific tomato recombinant inbred lines for fruit quality improvement. Metabolomics 2015, 11, 1416–1431. [Google Scholar] [CrossRef]
- Riedelsheimer, C.; Lisec, J.; Czedik-Eysenberg, A.; Sulpice, R.; Flis, A.; Grieder, C.; Altmann, T.; Stitt, M.; Willmitzer, L.; Melchinger, A.E. Genome-wide association mapping of leaf metabolic profiles for dissecting complex traits in maize. Proc. Natl. Acad. Sci. 2012, 109, 8872–8877. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riedelsheimer, C.; Czedik-Eysenberg, A.; Grieder, C.; Lisec, J.; Technow, F.; Sulpice, R.; Altmann, T.; Stitt, M.; Willmitzer, L.; Melchinger, A.E. Genomic and metabolic prediction of complex heterotic traits in hybrid maize. Nat. Genet. 2012, 44, 217–220. [Google Scholar] [CrossRef] [PubMed]
- Steinfath, M.; Strehmel, N.; Peters, R.; Schauer, N.; Groth, D.; Hummel, J.; Steup, M.; Selbig, J.; Kopka, J.; Geigenberger, P.; et al. Discovering plant metabolic biomarkers for phenotype prediction using an untargeted approach. Plant Biotechnol. J. 2010, 8, 900–911. [Google Scholar] [CrossRef] [PubMed]
- Keurentjes, J.J.B.; Fu, J.; de Vos, C.H.R.; Lommen, A.; Hall, R.D.; Bino, R.J.; van der Plas, L.H.W.; Jansen, R.C.; Vreugdenhil, D.; Koornneef, M. The genetics of plant metabolism. Nat. Genet. 2006, 38, 842–849. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alseekh, S.; Bermudez, L.; de Haro, L.A.; Fernie, A.R.; Carrari, F. Crop metabolomics: from diagnostics to assisted breeding. Metabolomics 2018, 14, 148. [Google Scholar] [CrossRef] [Green Version]
- Davies, H.V.; Shepherd, L.V.T.; Stewart, D.; Frank, T.; Röhlig, R.M.; Engel, K.-H. Metabolome variability in crop plant species – When, where, how much and so what? Regul. Toxicol. Pharmacol. 2010, 58, S54–S61. [Google Scholar] [CrossRef] [PubMed]
- Eskes, A.B.; Leroy, T. Coffee Selection and Breeding. In Coffee: Growing, Processing, Sustainable Production: A Guidebook for Growers, Processors, Traders and Researchers; John Wiley & Sons: Hoboken, NJ, USA, 2008; pp. 57–86. ISBN 978-3-527-61962-7. [Google Scholar]
- Singh, R.J. Genetic Resources, Chromosome Engineering, and Crop Improvement: Medicinal Plants; CRC Press: Boca Raton, FL, USA, 2012; ISBN 978-1-4200-7384-3. [Google Scholar]
- Sunarharum, W.B.; Williams, D.J.; Smyth, H.E. Complexity of coffee flavor: A compositional and sensory perspective. Food Res. Int. 2014, 62, 315–325. [Google Scholar] [CrossRef]
- Vvan der Vossen, H.; Bertrand, B.; Charrier, A. Next generation variety development for sustainable production of arabica coffee (Coffea arabica L.): a review. Euphytica 2015, 204, 243–256. [Google Scholar] [CrossRef]
- Bertrand, B.; Etienne, H.; Cilas, C.; Charrier, A.; Baradat, P. Coffea arabica hybrid performance for yield, fertility and bean weight. Euphytica 2005, 141, 255–262. [Google Scholar] [CrossRef]
- Costa Freitas, A.M.; Mosca, A.I. Coffee geographic origin — an aid to coffee differentiation. Food Res. Int. 1999, 32, 565–573. [Google Scholar] [CrossRef]
- Bertrand, B.; Vaast, P.; Alpizar, E.; Etienne, H.; Davrieux, F.; Charmetant, P. Comparison of bean biochemical composition and beverage quality of Arabica hybrids involving Sudanese-Ethiopian origins with traditional varieties at various elevations in Central America. Tree Physiol. 2006, 26, 1239–1248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bosselmann, A.S.; Dons, K.; Oberthur, T.; Olsen, C.S.; Ræbild, A.; Usma, H. The influence of shade trees on coffee quality in small holder coffee agroforestry systems in Southern Colombia. Agric. Ecosyst. Environ. 2009, 129, 253–260. [Google Scholar] [CrossRef]
- Bhumiratana, N.; Adhikari, K.; Chambers IV, E. Evolution of sensory aroma attributes from coffee beans to brewed coffee. LWT - Food Sci. Technol. 2011, 44, 2185–2192. [Google Scholar] [CrossRef] [Green Version]
- Poltronieri, Y.; Martinez, H.E.P.; Cecon, P.R. Effect of zinc and its form of supply on production and quality of coffee beans. J. Sci. Food Agric. 2011, 91, 2431–2436. [Google Scholar] [CrossRef]
- Buffo, R.A.; Cardelli-Freire, C. Coffee flavour: an overview. Flavour Fragr. J. 2004, 19, 99–104. [Google Scholar] [CrossRef]
- Viani, R.; Illy, A. Espresso Coffee: The Science of Quality, 2nd ed.; Elsevier: Amsterdam, The Netherlands, 2005; ISBN 978-0-12-370371-2. [Google Scholar]
- Baggenstoss, J.; Poisson, L.; Kaegi, R.; Perren, R.; Escher, F. Coffee Roasting and Aroma Formation: Application of Different Time−Temperature Conditions. J. Agric. Food Chem. 2008, 56, 5836–5846. [Google Scholar] [CrossRef]
- Ribeiro, J.S.; Augusto, F.; Salva, T.J.G.; Thomaziello, R.A.; Ferreira, M.M.C. Prediction of sensory properties of Brazilian Arabica roasted coffees by headspace solid phase microextraction-gas chromatography and partial least squares. Anal. Chim. Acta 2009, 634, 172–179. [Google Scholar] [CrossRef]
- Zhou, L.; Khalil, A.; Bindler, F.; Zhao, M.; Marcic, C.; Ennahar, S.; Marchioni, E. Effect of heat treatment on the content of individual phospholipids in coffee beans. Food Chem. 2013, 141, 3846–3850. [Google Scholar] [CrossRef]
- Lang, R.; Klade, S.; Beusch, A.; Dunkel, A.; Hofmann, T. Mozambioside Is an Arabica-Specific Bitter-Tasting Furokaurane Glucoside in Coffee Beans. J. Agric. Food Chem. 2015, 63, 10492–10499. [Google Scholar] [CrossRef]
- Iwasa, K.; Setoyama, D.; Shimizu, H.; Seta, H.; Fujimura, Y.; Miura, D.; Wariishi, H.; Nagai, C.; Nakahara, K. Identification of 3-Methylbutanoyl Glycosides in Green Coffea arabica Beans as Causative Determinants for the Quality of Coffee Flavors. J. Agric. Food Chem. 2015, 63, 3742–3751. [Google Scholar] [CrossRef]
- Stadler, R.H.; Varga, N.; Hau, J.; Vera, F.A.; Welti, D.H. Alkylpyridiniums. 1. Formation in Model Systems via Thermal Degradation of Trigonelline. J. Agric. Food Chem. 2002, 50, 1192–1199. [Google Scholar] [CrossRef]
- Bicho, N.C.; Leitão, A.E.; Ramalho, J.C.; Lidon, F.C. Identification of chemical clusters discriminators of the roast degree in Arabica and Robusta coffee beans. Eur. Food Res. Technol. 2011, 233, 303–311. [Google Scholar] [CrossRef]
- Los Santos-Briones, D.; Hernández-Sotomayor, S.M. Coffee biotechnology. Braz. J. Plant Physiol. 2006, 18, 217–227. [Google Scholar] [CrossRef]
- Mishra, M.K.; Slater, A. Recent Advances in the Genetic Transformation of Coffee. Biotechnol. Res. Int. 2012, 2012, 1–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schauer, N.; Semel, Y.; Balbo, I.; Steinfath, M.; Repsilber, D.; Selbig, J.; Pleban, T.; Zamir, D.; Fernie, A.R. Mode of Inheritance of Primary Metabolic Traits in Tomato. Plant Cell Online 2008, 20, 509–523. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toubiana, D.; Semel, Y.; Tohge, T.; Beleggia, R.; Cattivelli, L.; Rosental, L.; Nikoloski, Z.; Zamir, D.; Fernie, A.R.; Fait, A. Metabolic Profiling of a Mapping Population Exposes New Insights in the Regulation of Seed Metabolism and Seed, Fruit, and Plant Relations. PLoS Genet. 2012, 8, e1002612. [Google Scholar] [CrossRef] [PubMed]
- Alseekh, S.; Tohge, T.; Wendenberg, R.; Scossa, F.; Omranian, N.; Li, J.; Kleessen, S.; Giavalisco, P.; Pleban, T.; Mueller-Roeber, B.; et al. Identification and Mode of Inheritance of Quantitative Trait Loci for Secondary Metabolite Abundance in Tomato. Plant Cell 2015, 27, 485–512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stewart, D.; McDougall, G.J.; Sungurtas, J.; Verrall, S.; Graham, J.; Martinussen, I. Metabolomic approach to identifying bioactive compounds in berries: advances toward fruit nutritional enhancement. Mol. Nutr. Food Res. 2007, 51, 645–651. [Google Scholar] [CrossRef]
- Barbosa, J.N.; Borem, F.M.; Cirillo, M.A.; Malta, M.R.; Alvarenga, A.A.; Alves, H.M.R. Coffee Quality and Its Interactions with Environmental Factors in Minas Gerais, Brazil. J. Agric. Sci. 2012, 4, 181. [Google Scholar] [CrossRef]
- Bertrand, B.; Boulanger, R.; Dussert, S.; Ribeyre, F.; Berthiot, L.; Descroix, F.; Joët, T. Climatic factors directly impact the volatile organic compound fingerprint in green Arabica coffee bean as well as coffee beverage quality. Food Chem. 2012, 135, 2575–2583. [Google Scholar] [CrossRef]
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Orešič, M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinformatics 2010, 11, 395. [Google Scholar] [CrossRef] [PubMed]
- Gowda, H.; Ivanisevic, J.; Johnson, C.H.; Kurczy, M.E.; Benton, H.P.; Rinehart, D.; Nguyen, T.; Ray, J.; Kuehl, J.; Arevalo, B.; et al. Interactive XCMS Online: Simplifying Advanced Metabolomic Data Processing and Subsequent Statistical Analyses. Anal. Chem. 2014, 86, 6931–6939. [Google Scholar] [CrossRef] [PubMed]
- Winkler, R. SpiderMass: Semantic database creation and tripartite metabolite identification strategy. J. Mass Spectrom. 2015, 50, 538–541. [Google Scholar] [CrossRef] [PubMed]







| Number | Mother Plant ID | Sib 1 | Sib 2 | Number | Mother Plant ID | Sib 1 | Sib 2 |
|---|---|---|---|---|---|---|---|
| 1 | MP196 | 196-1 | 196-3 | 21 | MP349 | 349-2 | 349-3 |
| 2 | MP203 | 203-1 | 203-2 | 22 | MP360 | 360-3 | 360-4 |
| 3 | MP205 | 205-1 | 205-2 | 23 | MP390 | 390-1 | 390-3 |
| 4 | MP212 | 212-1 | 212-2 | 24 | MP408 | 408-1 | 408-4 |
| 5 | MP292 | 292-3 | 292-4 | 25 | MP416 | 416-1 | 416-3 |
| 6 | MP295 | 295-2 | 295-3 | 26 | MP418 | 418-3 | 418-4 |
| 7 | MP312 | 312-3 | 312-4 | 27 | MP419 | 419-2 | 419-4 |
| 8 | MP315 | 315-1 | 315-2 | 28 | MP420 | 420-1 | 420-4 |
| 9 | MP316 | 316-1 | 316-2 | 29 | MP430 | 430-2 | 430-3 |
| 10 | MP318 | 318-2 | 318-3 | 30 | MP432 | 432-1 | 432-2 |
| 11 | MP322 | 322-3 | 322-4 | 31 | MP433 | 433-2 | 433-3 |
| 12 | MP323 | 323-1 | 323-4 | 32 | MP434 | 434-2 | 434-4 |
| 13 | MP328 | 328-3 | 328-4 | 33 | MP442 | 442-2 | 442-3 |
| 14 | MP333 | 333-1 | 333-4 | 34 | MP443 | 443-3 | 443-4 |
| 15 | MP336 | 336-1 | 336-2 | 35 | MP449 | 449-1 | 449-3 |
| 16 | MP337 | 337-3 | 337-4 | 36 | MP450 | 450-3 | 450-4 |
| 17 | MP339 | 339-1 | 339-2 | 37 | MP451 | 451-1 | 451-4 |
| 18 | MP340 | 340-1 | 340-3 | 38 | MP452 | 452-2 | 452-3 |
| 19 | MP343 | 343-1 | 343-3 | 39 | MP453 | 453-2 | 453-3 |
| 20 | MP346 | 346-1 | 346-4 | 40 | MP454 | 454-2 | 454-4 |
| Sample | Aromatic Intensity | Aromatic Quality | Flavor | Astringency | Aftertaste | Acidity | Bitterness | Body | Sourness | Global Preference |
|---|---|---|---|---|---|---|---|---|---|---|
| MP302 | 3.27 ± 0.52 | 3.09 ± 0.33 | 2.73 ± 0.39 | 2.45 ± 0.49 | 3.27 ± 0.39 | 0.64 ± 0.46 | 3.45 ± 0.49 | 2.73 ± 0.52 | 0.27 ± 0.39 | 2.82 ± 0.29 |
| MP307 | 2.14 ± 0.33 | 1.86 ± 0.47 | 2.23 ± 0.41 | 2.05 ± 0.33 | 2.27 ± 0.38 | 0.55 ± 0.33 | 3.50 ± 0.54 | 2.23 ± 0.61 | 0.36 ± 0.46 | 2.14 ± 0.24 |
| MP309 | 3.18 ± 0.44 | 3.45 ± 0.49 | 3.09 ± 0.33 | 2.82 ± 0.29 | 3.00 ± 0.18 | 0.45 ± 0.49 | 2.82 ± 0.29 | 2.82 ± 0.29 | 0.55 ± 0.50 | 3.09 ± 0.16 |
| MP324 | 2.55 ± 0.49 | 2.64 ± 0.46 | 2.18 ± 0.29 | 2.45 ± 0.49 | 2.55 ± 0.67 | 0.55 ± 0.49 | 3.18 ± 0.47 | 2.36 ± 0.46 | 0.45 ± 0.49 | 2.36 ± 0.46 |
| MP316 | 2.64 ± 0.46 | 2.45 ± 0.49 | 2.91 ± 0.33 | 2.36 ± 0.46 | 2.64 ± 0.57 | 0.64 ± 0.46 | 3.73 ± 0.39 | 2.55 ± 0.49 | 0.45 ± 0.49 | 2.91 ± 0.16 |
| MP318 | 2.82 ± 0.29 | 2.64 ± 0.46 | 2.36 ± 0.46 | 3.09 ± 0.16 | 3.00 ± 0.18 | 0.55 ± 0.49 | 3.55 ± 0.49 | 2.73 ± 0.39 | 0.45 ± 0.49 | 2.55 ± 0.49 |
| MP323 | 3.00 ± 0.18 | 3.09 ± 0.16 | 3.09 ± 0.33 | 1.73 ± 0.39 | 2.64 ± 0.46 | 0.36 ± 0.46 | 3.55 ± 0.49 | 2.36 ± 0.57 | 0.18 ± 0.29 | 3.09 ± 0.33 |
| MP346 | 2.27 ± 0.39 | 2.09 ± 0.16 | 2.00 ± 0.36 | 2.09 ± 0.16 | 2.73 ± 0.39 | 0.45 ± 0.50 | 3.45 ± 0.49 | 2.73 ± 0.39 | 1.00 ± 0.18 | 2.00 ± 0.18 |
| MP360 | 2.64 ± 0.52 | 2.36 ± 0.46 | 1.82 ± 0.29 | 2.18 ± 0.29 | 2.55 ± 0.49 | 1.27 ± 1.07 | 2.36 ± 0.82 | 2.45 ± 0.77 | 1.27 ± 0.39 | 1.82 ± 0.29 |
| MP408 | 3.27 ± 0.39 | 2.82 ± 0.29 | 2.36 ± 0.46 | 2.82 ± 0.29 | 3.00 ± 0.36 | 0.27 ± 0.39 | 3.36 ± 0.46 | 2.73 ± 0.39 | 0.36 ± 0.36 | 2.82 ± 0.29 |
| MP420 | 2.73 ± 0.39 | 3.00 ± 0.18 | 3.00 ± 0.36 | 2.55 ± 0.49 | 2.91 ± 0.33 | 1.18 ± 0.29 | 3.36 ± 0.46 | 2.36 ± 0.46 | 0.55 ± 0.49 | 2.45 ± 0.49 |
| MP453 | 2.45 ± 0.49 | 1.82 ± 0.29 | 1.82 ± 0.47 | 2.45 ± 0.49 | 2.73 ± 0.39 | 1.00 ± 0.18 | 3.09 ± 0.16 | 2.64 ± 0.46 | 0.55 ± 0.49 | 1.91 ± 0.16 |
| MP454 | 2.91 ± 0.33 | 2.00 ± 0.18 | 1.91 ± 0.17 | 2.73 ± 0.39 | 2.55 ± 0.49 | 0.73 ± 0.4 | 2.91 ± 0.33 | 2.82 ± 0.29 | 0.64 ± 0.46 | 1.91 ± 0.16 |
| M495 | 2.36 ± 0.46 | 1.45 ± 0.49 | 1.45 ± 0.49 | 1.55 ± 0.59 | 1.73± 0.39 | 0.29 ± 0.74 | 2.27 ± 0.39 | 2.27 ± 0.39 | 0.91 ± 0.33 | 1.73 ± 0.52 |
| M498 | 2.73 ± 0.39 | 1.91 ± 0.16 | 1.36 ± 0.46 | 2.27 ± 0.39 | 2.09 ± 0.16 | 3.00 ± 0.18 | 1.91 ± 0.33 | 2.27 ± 0.40 | 2.00 ± 0.36 | 0.45 ± 0.49 |
| M499 | 3.09 ± 0.33 | 1.82 ± 0.29 | 1.55 ± 0.49 | 2.18 ± 0.29 | 1.73 ± 0.39 | 2.09 ± 0.49 | 2.18 ± 0.29 | 2.55 ± 0.49 | 1.18 ± 0.30 | 1.18 ± 0.29 |
| M500 | 3.00 ± 0.25 | 2.38 ± 0.46 | 2.25 ± 0.38 | 2.50 ± 0.50 | 2.50 ± 0.50 | 3.38 ± 0.46 | 2.25 ± 0.37 | 2.63 ± 0.46 | 1.63 ± 0.47 | 1.75 ± 0.37 |
| M502 | 3.43 ± 0.48 | 2.29 ± 0.40 | 1.71 ± 0.40 | 1.43 ± 0.48 | 2.43 ± 0.48 | 3.14 ± 0.24 | 2.29 ± 0.40 | 1.86 ± 0.24 | 1.14 ± 0.24 | 1.43 ± 0.49 |
| M506 | 2.88 ± 0.22 | 1.25 ± 0.38 | 1.50 ± 0.50 | 2.00 ± 0.25 | 2.50 ± 0.50 | 3.88 ± 0.44 | 1.75 ± 0.38 | 2.00 ± 0.25 | 3.13 ± 0.44 | 0.75 ± 0.38 |
| M507 | 2.57 ± 0.49 | 1.29 ± 0.41 | 1.57 ± 0.49 | 2.29 ± 0.69 | 1.57 ± 0.49 | 4.00 ± 0.57 | 1.43 ± 0.61 | 1.71 ± 0.41 | 1.43 ± 0.49 | 0.43 ± 0.49 |
| M508 | 2.71 ± 0.26 | 2.18 ± 0.29 | 2.00 ± 0.18 | 1.71 ± 0.40 | 1.43 ± 0.49 | 3.86 ± 0.24 | 1.29 ± 0.40 | 2.14 ± 0.24 | 1.14 ± 0.24 | 1.00 ± 0.28 |
| Sensory Attribute | HR m/z | Adduct Ion | Name | Tissue | Heritability | SCC | p-Value |
|---|---|---|---|---|---|---|---|
| Acidity | 963.4134 | n.a. | Unknown | Fruits | 0.96 | 0.667 | 4.98 × 10-2 |
| Aftertaste | 940.2271 | [M+Na]+ | 2,4-Decadienoyl-CoA | Fruits | 0.881 | 0.754 | 1.88 × 10-2 |
| Aromatic intensity | 860.5079 | n.a. | Unknown | Fruits | 0.999 | 0.812 | 7.89 × 10-3 |
| Aromatic quality | 536.0019 | n.a. | Unknown | Fruits | 0.656 | 0.800 | 9.63 × 10-3 |
| Astringency | 601.1263 | [M+Na]+ | 2,3-trans-proanthocyanidin | Fruits | 0.957 | 0.833 | 5.27 × 10-3 |
| Bitterness | 734.4907 | n.a. | Unknown | Fruits | 0.817 | 0.740 | 2.27 × 10-2 |
| Body | 789.1658 | [M+H]+ | Delphinidin 3-O-glucosyl-5-O-caffeoylglucoside | Fruits | 0.949 | 0.690 | 3.98 × 10-2 |
| Flavor | 598.5406 | n.a. | Unknown | Fruits | 0.998 | 0.740 | 2.27 × 10-2 |
| Global preference | 791.4983 | [M+H]+ | Nonaprenyl diphosphate | Fruits | 0.984 | 0.728 | 2.61 × 10-2 |
| Sourness | 521.5618 | n.a. | Unknown | Fruits | 0.842 | 0.689 | 4.00 × 10-2 |
| Acidity | 473.3339 | [M+H]+ | 6-Deoxoteasterone | Leaf | 0.643 | 0.783 | 1.13 × 10-2 |
| Aftertaste | 695.4571 | [M+Na]+ | PA(16:0/18:2) | Leaf | 0.535 | 0.737 | 2.40 × 10-2 |
| Aromatic intensity | 713.317 | n.a. | Unknown | Leaf | 0.958 | 0.803 | 9.01 × 10-3 |
| Aromatic quality | 251.2095 | [M+Na]+ | Myristic acid | Leaf | 0.994 | 0.766 | 1.62 × 10-2 |
| Astringency | 967.7852 | [M]− | MGDG(23:0/26:0) | Leaf | 0.612 | 0.666 | 4.90 × 10-2 |
| *Bitterness | 473.3339 | [M+H]+ | 6-Deoxoteasterone | Leaf | 0.643 | −0.857 | 3.14 × 10-3 |
| *Body | 463.3451 | [M+Na]+ | 4,4-Dimethylzymosterol | Leaf | 0.891 | −0.689 | 3.98 × 10-2 |
| Flavor | 251.2095 | [M+Na]+ | Myristic acid | Leaf | 0.994 | 0.672 | 4.89 × 10-2 |
| Global preference | 433.1626 | [M+K]+ | Gibberellin A28 | Leaf | 0.996 | 0.677 | 4.48 × 10-2 |
| Sourness | 473.3339 | [M+H]+ | 6-Deoxoteasterone | Leaf | 0.643 | 0.773 | 1.47 × 10-2 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gamboa-Becerra, R.; Hernández-Hernández, M.C.; González-Ríos, Ó.; Suárez-Quiroz, M.L.; Gálvez-Ponce, E.; Ordaz-Ortiz, J.J.; Winkler, R. Metabolomic Markers for the Early Selection of Coffea canephora Plants with Desirable Cup Quality Traits. Metabolites 2019, 9, 214. https://doi.org/10.3390/metabo9100214
Gamboa-Becerra R, Hernández-Hernández MC, González-Ríos Ó, Suárez-Quiroz ML, Gálvez-Ponce E, Ordaz-Ortiz JJ, Winkler R. Metabolomic Markers for the Early Selection of Coffea canephora Plants with Desirable Cup Quality Traits. Metabolites. 2019; 9(10):214. https://doi.org/10.3390/metabo9100214
Chicago/Turabian StyleGamboa-Becerra, Roberto, María Cecilia Hernández-Hernández, Óscar González-Ríos, Mirna L. Suárez-Quiroz, Eligio Gálvez-Ponce, José Juan Ordaz-Ortiz, and Robert Winkler. 2019. "Metabolomic Markers for the Early Selection of Coffea canephora Plants with Desirable Cup Quality Traits" Metabolites 9, no. 10: 214. https://doi.org/10.3390/metabo9100214
APA StyleGamboa-Becerra, R., Hernández-Hernández, M. C., González-Ríos, Ó., Suárez-Quiroz, M. L., Gálvez-Ponce, E., Ordaz-Ortiz, J. J., & Winkler, R. (2019). Metabolomic Markers for the Early Selection of Coffea canephora Plants with Desirable Cup Quality Traits. Metabolites, 9(10), 214. https://doi.org/10.3390/metabo9100214