Heat Stress-Induced Metabolic Remodeling in Saccharomyces cerevisiae
Abstract
:1. Introduction
2. Results and Discussion
2.1. Heat Stress-Induced Metabolic Responses Are Compartment-Specific
2.2. Lipid Metabolism
2.3. Purine and Pyrimidine Metabolism
2.4. Remodeling of Arginine Metabolism Upon Heat Stress
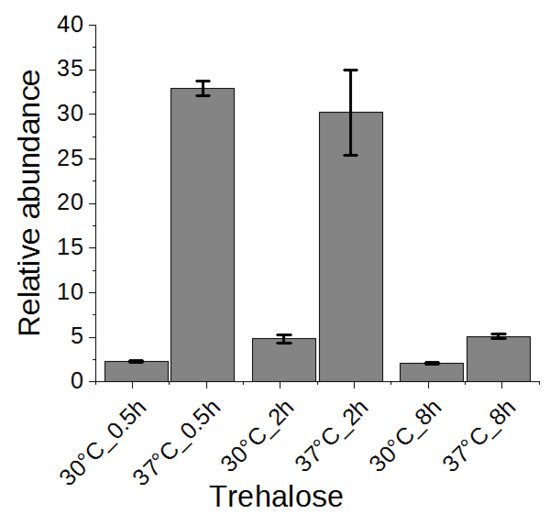
2.5. Trehalose
3. Conclusions
4. Materials and Methods
4.1. Yeast Culture
4.2. Heat Stress Treatment
4.3. Mitochondria Isolation
4.4. Metabolite Extraction, Metabolomics Analysis, and Data Evaluation
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Boy-Marcotte, E.; Lagniel, G.; Perrot, M.; Bussereau, F.; Boudsocq, A.; Jacquet, M.; Labarre, J. The heat shock response in yeast: Differential regulations and contributions of the Msn2p/Msn4p and Hsf1p regulons. Mol. Microbiol. 1999, 33, 274–283. [Google Scholar] [CrossRef] [PubMed]
- Causton, H.C.; Ren, B.; Koh, S.S.; Harbison, C.T.; Kanin, E.; Jennings, E.G.; Lee, T.I.; True, H.L.; Lander, E.S.; Young, R.A. Remodeling of yeast genome expression in response to environmental changes. Mol. Biol. Cell 2001, 12, 323–337. [Google Scholar] [CrossRef] [PubMed]
- Nagaraj, N.; Kulak, N.A.; Cox, J.; Neuhauser, N.; Mayr, K.; Hoerning, O.; Vorm, O.; Mann, M. System-wide perturbation analysis with nearly complete coverage of the yeast proteome by single-shot ultra HPLC runs on a bench top Orbitrap. Mol. Cell. Proteom. 2012, 11, M111.013722. [Google Scholar] [CrossRef] [PubMed]
- Strassburg, K.; Walther, D.; Takahashi, H.; Kanaya, S.; Kopka, J. Dynamic transcriptional and metabolic responses in yeast adapting to temperature stress. OMICS 2010, 14, 249–259. [Google Scholar] [CrossRef]
- Puig-Castellví, F.; Alfonso, I.; Piña, B.; Tauler, R. A quantitative 1H NMR approach for evaluating the metabolic response of Saccharomyces cerevisiae to mild heat stress. Metabolomics 2015, 11, 1612–1625. [Google Scholar] [CrossRef] [Green Version]
- Rikhvanov, E.G.; Lukina, E.A.; Varakina, N.N.; Rusaleva, T.M.; Gamburg, K.Z.; Knorre, D.A.; Borovskii, G.B.; Voinikov, V.K. Mitochondria as a critical element of heat shock response in yeasts with different types of energy metabolism. Russ. J. Plant Physiol. 2006, 53, 615–621. [Google Scholar] [CrossRef]
- Schmitt, M.; Neupert, W.; Langer, T. The molecular chaperone Hsp78 confers compartment-specific thermotolerance to mitochondria. J. Cell Biol. 1996, 134, 1375–1386. [Google Scholar] [CrossRef]
- Fiehn, O. Metabolite profiling in Arabidopsis. Methods Mol. Biol. 2006, 323, 439–447. [Google Scholar] [CrossRef]
- Pan, D.; Kather, M.; Willmann, L.; Schlimpert, M.; Bauer, C.; Lagies, S.; Schmidtkunz, K.; Eisenhardt, S.U.; Jung, M.; Gunther, S.; et al. Metabolic Response to XD14 Treatment in Human Breast Cancer Cell Line MCF-7. Int. J. Mol. Sci. 2016, 17, 1772. [Google Scholar] [CrossRef]
- Jain, M.; Nilsson, R.; Sharma, S.; Madhusudhan, N.; Kitami, T.; Souza, A.L.; Kafri, R.; Kirschner, M.W.; Clish, C.B.; Mootha, V.K. Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science 2012, 336, 1040–1044. [Google Scholar] [CrossRef]
- Chen, W.W.; Freinkman, E.; Wang, T.; Birsoy, K.; Sabatini, D.M. Absolute Quantification of Matrix Metabolites Reveals the Dynamics of Mitochondrial Metabolism. Cell 2016, 166, 1324–1337.e11. [Google Scholar] [CrossRef] [PubMed]
- Matuszczyk, J.-C.; Teleki, A.; Pfizenmaier, J.; Takors, R. Compartment-specific metabolomics for CHO reveals that ATP pools in mitochondria are much lower than in cytosol. Biotechnol. J. 2015, 10, 1639–1650. [Google Scholar] [CrossRef] [PubMed]
- Pan, D.; Lindau, C.; Lagies, S.; Wiedemann, N.; Kammerer, B. Metabolic profiling of isolated mitochondria and cytoplasm reveals compartment-specific metabolic responses. Metabolomics 2018, 14, 59. [Google Scholar] [CrossRef] [PubMed]
- Morgenstern, M.; Stiller, S.B.; Lübbert, P.; Peikert, C.D.; Dannenmaier, S.; Drepper, F.; Weill, U.; Höß, P.; Feuerstein, R.; Gebert, M.; et al. Definition of a High-Confidence Mitochondrial Proteome at Quantitative Scale. Cell Rep. 2017, 19, 2836–2852. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baker Brachmann, C.; Davies, A.; Cost, G.J.; Caputo, E.; Li, J.; Hieter, P.; Boeke, J.D. Designer deletion strains derived from Saccharomyces cerevisiae S288C: A useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 1998, 14, 115–132. [Google Scholar] [CrossRef]
- Ding, J.; Huang, X.; Zhang, L.; Zhao, N.; Yang, D.; Zhang, K. Tolerance and stress response to ethanol in the yeast Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2009, 85, 253–263. [Google Scholar] [CrossRef]
- Swan, T.M.; Watson, K. Stress tolerance in a yeast sterol auxotroph: Role of ergosterol, heat shock proteins and trehalose. FEMS Microbiol. Lett. 1998, 169, 191–197. [Google Scholar] [CrossRef]
- Dickson, R.C. Sphingolipid functions in Saccharomyces cerevisiae: Comparison to mammals. Annu. Rev. Biochem. 1998, 67, 27–48. [Google Scholar] [CrossRef]
- Cowart, L.A.; Obeid, L.M. Yeast sphingolipids: Recent developments in understanding biosynthesis, regulation, and function. Biochim. Biophys. Acta 2007, 1771, 421–431. [Google Scholar] [CrossRef] [Green Version]
- Kihara, A. Very long-chain fatty acids: Elongation, physiology and related disorders. J. Biochem. 2012, 152, 387–395. [Google Scholar] [CrossRef]
- Jenkins, G.M.; Hannun, Y.A. Role for de novo sphingoid base biosynthesis in the heat-induced transient cell cycle arrest of Saccharomyces cerevisiae. J. Biol. Chem. 2001, 276, 8574–8581. [Google Scholar] [CrossRef] [PubMed]
- Kurtz, J.-E.; Exinger, F.; Erbs, P.; Jund, R. The URH1 uridine ribohydrolase of Saccharomyces cerevisiae. Curr. Genet. 2002, 41, 132–141. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, A.; Kawahara, N.; Takagi, H. The flavoprotein Tah18-dependent NO synthesis confers high-temperature stress tolerance on yeast cells. Biochem. Biophys. Res. Commun. 2013, 430, 137–143. [Google Scholar] [CrossRef] [PubMed]
- Almeida, B.; Buttner, S.; Ohlmeier, S.; Silva, A.; Mesquita, A.; Sampaio-Marques, B.; Osório, N.S.; Kollau, A.; Mayer, B.; Leão, C.; et al. NO-mediated apoptosis in yeast. J. Cell Sci. 2007, 120, 3279–3288. [Google Scholar] [CrossRef] [Green Version]
- Nasuno, R.; Aitoku, M.; Manago, Y.; Nishimura, A.; Sasano, Y.; Takagi, H. Nitric oxide-mediated antioxidative mechanism in yeast through the activation of the transcription factor Mac1. PLoS ONE 2014, 9, e113788. [Google Scholar] [CrossRef]
- Astuti, R.I.; Nasuno, R.; Takagi, H. Nitric oxide signaling in yeast. Appl. Microbiol. Biotechnol. 2016, 100, 9483–9497. [Google Scholar] [CrossRef]
- Hamasaki-Katagiri, N.; Tabor, C.W.; Tabor, H. Spermidine biosynthesis in Saccharomyces cerevisae: Polyamine requirement of a null mutant of the SPE3 gene (spermidine synthase). Gene 1997, 187, 35–43. [Google Scholar] [CrossRef]
- Desiderio, M.A.; Dansi, P.; Tacchini, L.; Bernelli-Zazzera, A. Influence of polyamines on DNA binding of heat shock and activator protein 1 transcription factors induced by heat shock. FEBS Lett. 1999, 455, 149–153. [Google Scholar] [CrossRef]
- Sagor, G.H.M.; Berberich, T.; Takahashi, Y.; Niitsu, M.; Kusano, T. The polyamine spermine protects Arabidopsis from heat stress-induced damage by increasing expression of heat shock-related genes. Transgenic Res. 2013, 22, 595–605. [Google Scholar] [CrossRef]
- Singer, M.A.; Lindquist, S. Thermotolerance in Saccharomyces cerevisiae: The Yin and Yang of trehalose. Trends Biotechnol. 1998, 16, 460–468. [Google Scholar] [CrossRef]
- Eleutherio, E.; Panek, A.; de Mesquita, J.F.; Trevisol, E.; Magalhães, R. Revisiting yeast trehalose metabolism. Curr. Genet. 2015, 61, 263–274. [Google Scholar] [CrossRef] [PubMed]
- Gibney, P.A.; Schieler, A.; Chen, J.C.; Rabinowitz, J.D.; Botstein, D. Characterizing the in vivo role of trehalose in Saccharomyces cerevisiae using the AGT1 transporter. Proc. Natl. Acad. Sci. USA 2015, 112, 6116–6121. [Google Scholar] [CrossRef] [PubMed]
- Domitrovic, T.; Palhano, F.; Barjafidalgo, C.; Defreitas, M.; Orlando, M.; Fernandes, P. Role of nitric oxide in the response of cells to heat shock and high hydrostatic pressure. FEMS Yeast Res. 2003, 3, 341–346. [Google Scholar] [CrossRef]
- Ippolito, D.L.; Lewis, J.A.; Yu, C.; Leon, L.R.; Stallings, J.D. Alteration in circulating metabolites during and after heat stress in the conscious rat: Potential biomarkers of exposure and organ-specific injury. BMC Physiol. 2014, 14, 14. [Google Scholar] [CrossRef]
- Erez, A.; Nagamani, S.C.S.; Shchelochkov, O.A.; Premkumar, M.H.; Campeau, P.M.; Chen, Y.; Garg, H.K.; Li, L.; Mian, A.; Bertin, T.K.; et al. Requirement of argininosuccinate lyase for systemic nitric oxide production. Nat. Med. 2011, 17, 1619–1626. [Google Scholar] [CrossRef] [Green Version]
- Yoshikawa, Y.; Nasuno, R.; Kawahara, N.; Nishimura, A.; Watanabe, D.; Takagi, H. Regulatory mechanism of the flavoprotein Tah18-dependent nitric oxide synthesis and cell death in yeast. Nitric Oxide 2016, 57, 85–91. [Google Scholar] [CrossRef]
- Vernis, L.; Facca, C.; Delagoutte, E.; Soler, N.; Chanet, R.; Guiard, B.; Faye, G.; Baldacci, G. A newly identified essential complex, Dre2-Tah18, controls mitochondria integrity and cell death after oxidative stress in yeast. PLoS ONE 2009, 4, e4376. [Google Scholar] [CrossRef]
- Hartl, J.; Kiefer, P.; Meyer, F.; Vorholt, J.A. Longevity of major coenzymes allows minimal de novo synthesis in microorganisms. Nat. Microbiol. 2017, 2, 17073. [Google Scholar] [CrossRef]
- Lagies, S.; Pichler, R.; Kaminski, M.M.; Schlimpert, M.; Walz, G.; Lienkamp, S.S.; Kammerer, B. Metabolic characterization of directly reprogrammed renal tubular epithelial cells (iRECs). Sci. Rep. 2018, 8, 3878. [Google Scholar] [CrossRef]







© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pan, D.; Wiedemann, N.; Kammerer, B. Heat Stress-Induced Metabolic Remodeling in Saccharomyces cerevisiae. Metabolites 2019, 9, 266. https://doi.org/10.3390/metabo9110266
Pan D, Wiedemann N, Kammerer B. Heat Stress-Induced Metabolic Remodeling in Saccharomyces cerevisiae. Metabolites. 2019; 9(11):266. https://doi.org/10.3390/metabo9110266
Chicago/Turabian StylePan, Daqiang, Nils Wiedemann, and Bernd Kammerer. 2019. "Heat Stress-Induced Metabolic Remodeling in Saccharomyces cerevisiae" Metabolites 9, no. 11: 266. https://doi.org/10.3390/metabo9110266
APA StylePan, D., Wiedemann, N., & Kammerer, B. (2019). Heat Stress-Induced Metabolic Remodeling in Saccharomyces cerevisiae. Metabolites, 9(11), 266. https://doi.org/10.3390/metabo9110266