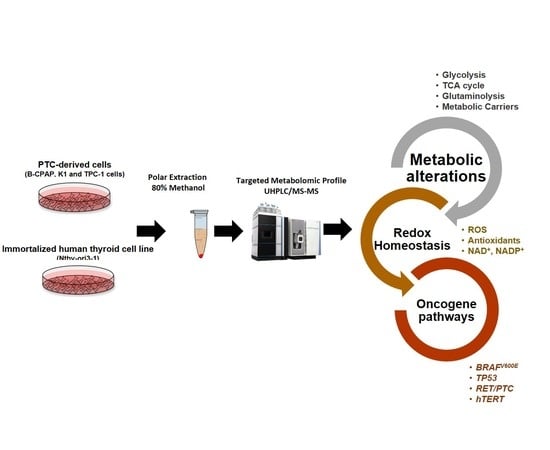
Crosstalk between Metabolic Alterations and Altered Redox Balance in PTC-Derived Cell Lines
Abstract
:1. Introduction
2. Results
2.1. Metabolomic Profiles of PTC-Derived Cell Lines
2.2. Expression of GLUT1 and MCT4 Transporters and Glucose Uptake Results
2.3. Redox Alterations in PTC Cells
2.4. Expression of Glutamate Cysteine Ligase and Glutaminase 1
3. Discussion
3.1. Metabolic Alterations in PTC Cells
3.2. Redox Alterations in PTC Cells
4. Conclusions
5. Material and Methods
5.1. Cell Culture
5.2. Aqueous Metabolites Extraction
5.3. Ultra High Performance Liquid Chromatography-Tandem Mass Spectrometry
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Ward, P.S.; Thompson, C.B. Metabolic Reprogramming: A Cancer Hallmark Even Warburg Did Not Anticipate. Cancer Cell 2012, 21, 297–308. [Google Scholar] [CrossRef] [PubMed]
- Locasale, J.W.; Cantley, L.C. Altered metabolism in cancer. BMC Biol. 2010, 25, 8–88. [Google Scholar] [CrossRef] [PubMed]
- Locasale, J.W.; Cantley, L.C.; Vander Heiden, M.G. Cancer’s insatiable appetite. Nat. Biotechnol. 2009, 27, 916–917. [Google Scholar] [CrossRef] [PubMed]
- Teicher, B.A.; Linehan, W.M.; Helman, L.J. Targeting Cancer Metabolism. Clin. Cancer Res. 2012, 18, 5537–5545. [Google Scholar] [CrossRef] [PubMed]
- Jackson, A.L.; Loeb, L.A. The contribution of endogenous sources of DNA damage to the multiple mutations in cancer. Mutat. Res. 2001, 477, 7–21. [Google Scholar] [CrossRef]
- Schafer, Z.T.; Grassian, A.R.; Song, L.; Jiang, Z.; Gerhart-Hines, Z.; Irie, H.Y.; Gao, S.; Puigserver, P.; Brugge, J.S. Antioxidant and oncogene rescue of metabolic defects caused by loss of matrix attachment. Nature 2009, 461, 109–113. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Hyde, A.S.; Simpson, M.A.; Barycki, J.J. Emerging regulatory paradigms in glutathione metabolism. Adv. Cancer Res. 2014, 122, 69–101. [Google Scholar] [CrossRef]
- Cairns, R.A.; Harris, I.S.; Mak, T.W. Regulation of cancer cell metabolism. Nat. Rev. Cancer 2011, 11, 85–95. [Google Scholar] [CrossRef] [PubMed]
- Xing, M. BRAF mutation in thyroid cancer. Endocr. Relat. Cancer 2005, 12, 245–262. [Google Scholar] [CrossRef] [PubMed]
- Giordano, T.J. Follicular cell thyroid neoplasia: Insights from genomics and The Cancer Genome Atlas research network. Curr. Opin. Oncol. 2016, 28, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Nikiforov, Y.E.; Nikiforova, M.N. Molecular genetics and diagnosis of thyroid cancer. Nat. Rev. Endocrinol. 2011, 7, 569–580. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Qu, S.; Liu, R.; Sheng, C.; Shi, X.; Zhu, G.; Murugan, A.; Guan, H.; Yu, H.; Wang, Y.; et al. TERT promoter mutations and their association with BRAF V600E mutation and aggressive clinicopathological characteristics of thyroid cancer. J. Clin. Endocrinol. Metab. 2014, 99, E1130–E1136. [Google Scholar] [CrossRef] [PubMed]
- Asa, S.L. The evolution of differentiated thyroid cancer. Pathology 2017, 49, 229–237. [Google Scholar] [CrossRef] [PubMed]
- Liesenfeld, D.B.; Habermann, N.; Owen, R.W.; Scalbert, A.; Ulrich, C.M. Review of Mass Spectrometry-Based Metabolomics in Cancer Research. Cancer Epidemiol. Biomark. Prev. 2013, 22, 2182–2201. [Google Scholar] [CrossRef] [PubMed]
- Caria, P.; Pillai, R.; Dettori, T.; Frau, D.V.; Zavattari, P.; Riva, G.; Romano, G.; Pani, F.; Bentivegna, A.; Giovannoni, R.; et al. Thyrospheres from B-CPAP Cell Line with BRAF and TERT Promoter Mutations have Different Functional and Molecular Features than Parental Cells. J. Cancer 2017, 8, 1629–1639. [Google Scholar] [CrossRef] [PubMed]
- Nagarajah, J.; Ho, A.L.; Tuttle, R.M.; Webwe, W.A.; Grewal, R.K. Correlation of BRAFV600E mutation and glucose metabolism in thyroid cancer patients: An 18F-FDG PET study. J. Nucl. Med. 2015, 56, 662–667. [Google Scholar] [CrossRef]
- Hutton, J.E.; Wang, X.; Zimmerman, L.J.; Slebos, R.J.; Trenary, I.A.; Young, J.D.; Li, M.; Liebler, D.C. Oncogenic KRAS and BRAF Drive Metabolic Reprogramming in Colorectal Cancer. Mol. Cell. Proteom. 2016, 15, 2924–2938. [Google Scholar] [CrossRef]
- Bensaad, K.; Tsuruta, A.; Selak, M.A.; Vidal, M.N.; Nakano, K.; Bartrons, R.; Gottlieb, E.; Vousden, K.H. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell 2006, 126, 107–120. [Google Scholar] [CrossRef]
- Ahn, C.S.; Metallo, C.M. Mitochondria as biosynthetic factories for cancer proliferation. Cancer Metab. 2015, 3, 1. [Google Scholar] [CrossRef]
- Chen, J.Q.; Russo, J. Dysregulation of glucose transport, glycolysis, TCA cycle and glutaminolysis by oncogenes and tumor suppressors in cancer cells. Biochim. Biophys. Acta 2012, 1826, 370–384. [Google Scholar] [CrossRef]
- Pavlova, N.N.; Thompson, C.B. The emerging hallmarks of cancer metabolism. Cell Metab. 2016, 23, 27–47. [Google Scholar] [CrossRef] [PubMed]
- Icard, P.; Poulain, L.; Licet, H. Understanding the central role of citric acid in the metabolism of cancer cells. Biochim. Biophys. Acta 2012, 1825, 111–116. [Google Scholar] [CrossRef] [PubMed]
- Feron, O. Pyruvate into lactic acid and back: From the Warburg effect to symbiotic energy fuel exchange in cancer cells. Radiother. Oncol. 2009, 92, 329–333. [Google Scholar] [CrossRef] [PubMed]
- Fuhler, G.M.; Eppinga, H.; Peppelenbosch, M.P. Fumaric acids and Cancer. Trends Mol. Med. 2017, 23, 3–5. [Google Scholar] [CrossRef] [PubMed]
- Brabletz, T.; Kalluri, R.; Nieto, M.S.; Weinberg, R.A. ETM in cancer. Nat. Rev. Cancer 2018, 18, 128–134. [Google Scholar] [CrossRef] [PubMed]
- Jóźwiak, P.; Krześlak, A.; Bryś, M.; Lipińska, A. Glucose-dependent glucose transporter 1 expression and its impact on viability of thyroid cancer cells. Oncol. Rep. 2015, 33, 913–920. [Google Scholar] [CrossRef] [PubMed]
- Jiang, B. Aerobic glycolysis and high level of lactic acid in cancer metabolism and microenvironment. Genes Dis. 2017, 4, 25–27. [Google Scholar] [CrossRef]
- Choi, S.Y.; Xue, H.; Wu, R.; Fazli, L.; Lin, D.; Collins, C.C.; Gleave, M.E.; Gout, P.W.; Wang, Y. The MCT4 Gene: A Novel, Potential Target for Therapy of Advanced Prostate Cancer. Clin. Cancer Res. 2016, 22, 2721–2733. [Google Scholar] [CrossRef]
- Lee, J.Y.; Lee, I.; Chang, W.J.; Ahn, S.M.; Lim, S.H.; Kim, H.S.; Yoo, K.H.; Jung, K.S.; Song, H.N.; Cho, J.H.; et al. MCT4 as a potential therapeutic target for metastatic gastric cancer with peritoneal carcinomatosis. Oncotarget 2016, 7, 43492–43503. [Google Scholar] [CrossRef]
- DeBerardinis, R.J.; Mancuso, A.; Daikhin, E.; Nissim, I.; Yudkoff, M.; Wehrli, S.; Thompson, C.B. Beyond aerobic glycolysis: Transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc. Natl. Acad. Sci. USA 2007, 104, 19345–19350. [Google Scholar] [CrossRef]
- Anderson, N.M.; Mucka, P.; Kern, J.G.; Feng, H. The emerging role and targetability if the TCA cycle in cancer metabolism. Protein Cell 2018, 9, 216–237. [Google Scholar] [CrossRef] [PubMed]
- Maddocks, O.D.; Vousden, K.H. Metabolic Regulation by p53. J. Mol. Med. (Berl.) 2011, 89, 237–245. [Google Scholar] [CrossRef]
- Gottlieb, E.; Vousden, K.H. P53 regulation of metabolic pathways. Cold Spring Harb. Perspect. Biol. 2012, 2, a001040. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.C.; Hevia, D.; Patchva, S.; Park, B.; Koh, W.; Aggarwal, B.B. Upsides and downsides of reactive oxygen species for cancer: The roles of reactive oxygen species in tumorigenesis, prevention, and therapy. Antioxid. Redox Signal. 2012, 16, 1295–1322. [Google Scholar] [CrossRef] [PubMed]
- Ying, W. NAD+/NADH and NADP+/NADPH in cellular functions and cell death: Regulation and biological consequences. Antioxid. Redox Signal. 2008, 10, 179–206. [Google Scholar] [CrossRef] [PubMed]
- Estrela, J.M.; Ortega, A.; Obrador, E. Glutathione in cancer biology and therapy. Crit. Rev. Clin. Lab. Sci. 2006, 43, 143–181. [Google Scholar] [CrossRef] [PubMed]
- Health Protection Agency Culture Collections. Available online: http://www.hpa.org.uk (accessed on 5 May 2011).
- Caria, P.; Tronci, L.; Dettori, T.; Murgia, F.; Santoru, M.L.; Griffin, J.L.; Vanni, R.; Atzori, L. Metabolomic Alterations in Thyrospheres and Adherent Parental Cells in Papillary Thyroid Carcinoma Cell Lines: A Pilot Study. Int. J. Mol. Sci. 2018, 27, 19. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.; Khan, M.I.; Iqbal, Z.; Shah, Y.; Ahmad, L.; Nazir, S.; Watson, D.G.; Khan, J.A.; Nasir, F.; Khan, A.; et al. A new HPLC method for the simultaneous determination of ascorbic acid and aminothiols in human plasma and erythrocytes using electrochemical detection. Talanta 2011, 84, 789–801. [Google Scholar] [CrossRef]
- Dinicola, S.; Mariggiò, M.A.; Morabito, C.; Guarnieri, S.; Cucina, A.; Pasqualato, A.; D’Anselmi, F.; Proietti, S.; Coluccia, P.; Bizzarri, M. Grape seed extract triggers apoptosis in Caco-2 human colon cancer cells through reactive oxygen species and calcium increase: Extracellular signal-regulated kinase involvement. Br. J. Nutr. 2013, 110, 797–809. [Google Scholar] [CrossRef]






| Cell Lines | Tissue Origin | hTERT | BRAFV600E | RET/PTC | TP53 |
|---|---|---|---|---|---|
| B-CPAP | PTC | C228T | GTG -> GAG * | Wt | GAC -> TAC |
| K1 | PTC | C228T | GTG -> GAG ** | Wt | wt |
| TPC-1 | PTC | C228T | wt | RET/PTC1 | wt |
| Nthy-ori 3-1 | NTT | wt | wt | wt | wt |
| Compound | RT | Precursorion (m/z) | Production (m/z) | Polarity | Column |
|---|---|---|---|---|---|
| 2/3 Phosphoglycerate | 4.02 | 185.000 | 97.065 | Negative | BEH amide |
| Acetyl-CoA | 3.65 | 810.225 | 303.049 | Positive | BEH amide |
| α-ketoglutarate | 3.35 | 145 | 101.123 | Negative | BEH amide |
| Citric acid | 1.32 | 191 | 111 | Negative | C18-pfp |
| Dihydroxyacetone phosphate | 3.88 | 169 | 97.143 | Negative | BEH amide |
| Glucose-6-phosphate/Fructose 6 phosphate | 4.05 | 259 | 97.077 | Negative | BEH amide |
| Fructose bisphosphate | 4.24 | 339 | 97.084 | Negative | BEH amide |
| Fumaric Acid | 3.48 | 115 | 71.172 | Negative | BEH amide |
| Glucose | 2 | 179 | 89 | Positive | BEH amide |
| Glutamic Acid | 0.74 | 148 | 84.15 | Positive | C18-pfp |
| Glutamine | 0.72 | 153.1 | 89.169 | Positive | C18-pfp |
| Glyceraldhehide-3-phosphate | 3.83 | 169.009 | 97 | Negative | BEH amide |
| Isocitric acid | 1.71 | 191 | 111 | Negative | C18-pfp |
| Lactic Acid | 2.5 | 89 | 43 | Negative | BEH amide |
| Malic Acid | 3.67 | 133 | 115.081 | Negative | BEH amide |
| NAD+ | 3.82 | 664.1 | 136.237 | Positive | BEH amide |
| NADP+ | 4.15 | 744.1 | 136.2 | Positive | BEH amide |
| Oxaloacetate | 3.34 | 131 | 87.084 | Negative | BEH amide |
| Phosphoenolpyruvate | 3.94 | 167 | 79.081 | Negative | BEH amide |
| Pyruvic acid | 2.75 | 87 | 43 | Negative | BEH amide |
| Succinic acid | 3.51 | 117 | 73 | Negative | BEH amide |
| Succynil-CoA | 3.92 | 868.1 | 361.214 | Positive | BEH amide |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tronci, L.; Caria, P.; Frau, D.V.; Liggi, S.; Piras, C.; Murgia, F.; Santoru, M.L.; Pibiri, M.; Deiana, M.; Griffin, J.L.; et al. Crosstalk between Metabolic Alterations and Altered Redox Balance in PTC-Derived Cell Lines. Metabolites 2019, 9, 23. https://doi.org/10.3390/metabo9020023
Tronci L, Caria P, Frau DV, Liggi S, Piras C, Murgia F, Santoru ML, Pibiri M, Deiana M, Griffin JL, et al. Crosstalk between Metabolic Alterations and Altered Redox Balance in PTC-Derived Cell Lines. Metabolites. 2019; 9(2):23. https://doi.org/10.3390/metabo9020023
Chicago/Turabian StyleTronci, Laura, Paola Caria, Daniela Virginia Frau, Sonia Liggi, Cristina Piras, Federica Murgia, Maria Laura Santoru, Monica Pibiri, Monica Deiana, Julian Leether Griffin, and et al. 2019. "Crosstalk between Metabolic Alterations and Altered Redox Balance in PTC-Derived Cell Lines" Metabolites 9, no. 2: 23. https://doi.org/10.3390/metabo9020023
APA StyleTronci, L., Caria, P., Frau, D. V., Liggi, S., Piras, C., Murgia, F., Santoru, M. L., Pibiri, M., Deiana, M., Griffin, J. L., Vanni, R., & Atzori, L. (2019). Crosstalk between Metabolic Alterations and Altered Redox Balance in PTC-Derived Cell Lines. Metabolites, 9(2), 23. https://doi.org/10.3390/metabo9020023