Translational Metabolomics: Current Challenges and Future Opportunities
Abstract
:1. Introduction
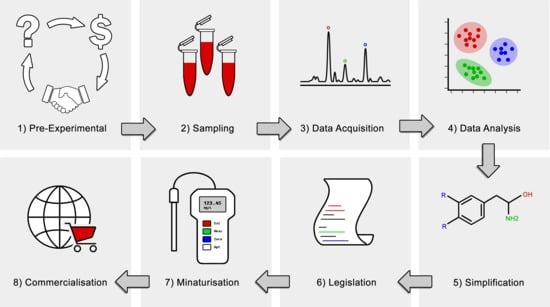
2. Translational Omics: Where Are We Now?
3. Translational Challenges in Metabolomics
3.1. Perceptions
3.2. Costs
3.3. Expertise
3.4. Data Acquisition
3.5. Metabolite Identification and Pathway Mapping
3.6. Quantification
4. Translational Opportunities in Metabolomics
4.1. Simplification
4.2. Commercialisation
4.3. Test Format and Miniaturisation
5. Clinical and Industrial Applications of Metabolomics
5.1. Biomarker Discovery and Development of Diagnostic Tools
5.2. Personalised Medicine and Nutrition
5.3. Drug Targets and Development
5.4. Industrial Applications
6. Recommendations
- (a)
- Methods for absolute quantification of metabolites (targeted and untargeted) using different analytical instrumentations should be improved. Thus far, most of the quantitative methods are targeted, therefore, some special attention needs to be in place for the development of untargeted quantitative metabolomics methods as this will be highly useful in biomarker discovery related work. However, we agree that targeted quantification of metabolites that is done well will be more useful than poorly done untargeted metabolite profiling.
- (b)
- More research on how to make miniaturised instruments, make them less expensive and accessible should be encouraged. This will ultimately allow anyone interested in metabolite measurement to undertake metabolomics studies. This can be carried out with the support from vendors involved in developing different analytical platforms through a mutual collaboration.
- (c)
- Automated data processing, development of user friendly software and databases should be encouraged, particularly efforts should be undertaken to develop more open source web-based data analysis platforms. This can make data interpretation more robust and will open up avenues for more translational metabolomics research.
- (d)
- If biomarker and drug target discovery is the main target of a metabolomics study, then all the guidelines provided by professional and regulatory bodies regarding better experimental design, data acquisition and validation should be carried out. Successful translation of new biomarkers only will be possible if the strategies and implementation pathways are considered since the beginning of a project.
- (e)
- The metabolomics community should work along with other omics communities to establish a better platform for multi-omics integration in order to gain overall insights on cellular processes. This will not only be helpful with translational opportunities, but also with acquiring funds from different governmental and industrial bodies.
- (f)
- It is extremely important to organise forums or symposiums where a cross talk among different professional bodies, government organizations, regularity and funding bodies can take place. Collaborative approach will definitely provide more translational opportunities for the omics community.
- (g)
- The metabolomics community should also encourage publishing their outcomes to journals, newspapers and social media to raise more social consciousness on personalised medicine and nutrition by providing more scientific evidence. It is already clear that “one glove fits all” does not work when it comes to translation of scientific results to clinics or industries.
7. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Dunn, W.B. Current trends and future requirements for the mass spectrometric investigation of microbial, mammalian and plant metabolomes. Phys. Biol. 2008, 5, 011001. [Google Scholar] [CrossRef] [PubMed]
- Trivedi, D.K.; Hollywood, K.A.; Goodacre, R. Metabolomics for the masses: The future of metabolomics in a personalized world. New Horiz. Transl. Med. 2017, 3, 294–305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Savolainen, O.; Fagerberg, B.; Vendelbo Lind, M.; Sandberg, A.-S.; Ross, A.B.; Bergström, G. Biomarkers for predicting type 2 diabetes development-Can metabolomics improve on existing biomarkers? PLoS ONE 2017, 12, e0177738. [Google Scholar] [CrossRef] [PubMed]
- Burton, C.; Ma, Y.F. Current Trends in Cancer Biottiarker Discovery Using Urinary Metabolomics: Achievements and New Challenges. Curr. Med. Chem. 2019, 26, 5–28. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.; Lu, H.; Lee, Y.H. Challenges and emergent solutions for LC-MS/MS based untargeted metabolomics in diseases. Mass Spectrom. Rev. 2018, 37, 772–792. [Google Scholar] [CrossRef] [PubMed]
- Beale, D.J.; Pinu, F.R.; Kouremenos, K.A.; Poojary, M.M.; Narayana, V.K.; Boughton, B.A.; Kanojia, K.; Dayalan, S.; Jones, O.A.H.; Dias, D.A. Review of recent developments in GC–MS approaches to metabolomics-based research. Metabolomics 2018, 14, 152. [Google Scholar] [CrossRef] [PubMed]
- Chong, J.; Soufan, O.; Caraus, I.; Xia, J.; Li, C.; Wishart, D.S.; Bourque, G.; Li, S. MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018, 46, W486–W494. [Google Scholar] [CrossRef]
- Djoumbou-Feunang, Y.; Pon, A.; Karu, N.; Zheng, J.; Li, C.; Arndt, D.; Gautam, M.; Allen, F.; Wishart, S.D. CFM-ID 3.0: Significantly Improved ESI-MS/MS Prediction and Compound Identification. Metabolites 2019, 9, 72. [Google Scholar] [CrossRef]
- Pinu, R.F. Grape and Wine Metabolomics to Develop New Insights Using Untargeted and Targeted Approaches. Fermentation 2018, 4, 92. [Google Scholar] [CrossRef]
- Oliver, S.G.; Winson, M.K.; Kell, D.B.; Baganz, F. Systematic functional analysis of the yeast genome. Trends Biotechnol. 1998, 16, 373–378. [Google Scholar] [CrossRef]
- Fiehn, O. Metabolomics—The link between genotypes and phenotypes. In Functional Genomics; Town, C., Ed.; Springer: Dordrecht, The Netherlands, 2002; pp. 155–171. [Google Scholar]
- Wishart, D.S. Emerging applications of metabolomics in drug discovery and precision medicine. Nat. Rev. Drug Discov. 2016, 15, 473. [Google Scholar] [CrossRef] [PubMed]
- Kell, D.B.; Oliver, S.G. The metabolome 18 years on: A concept comes of age. Metab. Off. J. Metab. Soc. 2016, 12, 148. [Google Scholar] [CrossRef] [PubMed]
- Beger, R.D.; Dunn, W.B.; Bandukwala, A.; Bethan, B.; Broadhurst, D.; Clish, C.B.; Dasari, S.; Derr, L.; Evans, A.; Fischer, S.; et al. Towards quality assurance and quality control in untargeted metabolomics studies. Metabolomics 2019, 15, 4. [Google Scholar] [CrossRef] [PubMed]
- Pinu, F.R. Metabolomics: Applications to Food Safety and Quality Research. In Microbial Metabolomics: Applications in Clinical, Environmental, and Industrial Microbiology; Beale, D.J., Kouremenos, K.A., Palombo, E.A., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 225–259. [Google Scholar]
- Skelton, D.M.; Ekman, D.R.; Martinović-Weigelt, D.; Ankley, G.T.; Villeneuve, D.L.; Teng, Q.; Collette, T.W. Metabolomics for in Situ Environmental Monitoring of Surface Waters Impacted by Contaminants from Both Point and Nonpoint Sources. Environ. Sci. Technol. 2014, 48, 2395–2403. [Google Scholar] [PubMed]
- Wishart, D.S. Metabolomics: Applications to food science and nutrition research. Trends Food Sci. Technol. 2008, 19, 482–493. [Google Scholar] [CrossRef]
- Kim, S.; Kim, J.; Yun, E.J.; Kim, K.H. Food metabolomics: From farm to human. Curr. Opin. Biotechnol. 2016, 37, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Pinu, F.R. Metabolomics—The new frontier in food safety and quality research. Food Res. Int. 2015, 72, 80–81. [Google Scholar] [CrossRef]
- Liu, J.; Semiz, S.; van der Lee, S.J.; van der Spek, A.; Verhoeven, A.; van Klinken, J.B.; Sijbrands, E.; Harms, A.C.; Hankemeier, T.; van Dijk, K.W.; et al. Metabolomics based markers predict type 2 diabetes in a 14-year follow-up study. Metab. Off. J. Metab. Soc. 2017, 13, 104. [Google Scholar] [CrossRef]
- Shah Svati, H.; Kraus William, E.; Newgard Christopher, B. Metabolomic Profiling for the Identification of Novel Biomarkers and Mechanisms Related to Common Cardiovascular Diseases. Circulation 2012, 126, 1110–1120. [Google Scholar] [CrossRef] [Green Version]
- Shajahan-Haq, A.N.; Cheema, M.S.; Clarke, R. Application of metabolomics in drug resistant breast cancer research. Metabolites 2015, 5, 100–118. [Google Scholar] [CrossRef]
- Mehta, K.Y.; Wu, H.-J.; Menon, S.S.; Fallah, Y.; Zhong, X.; Rizk, N.; Unger, K.; Mapstone, M.; Fiandaca, M.S.; Federoff, H.J.; et al. Metabolomic biomarkers of pancreatic cancer: A meta-analysis study. Oncotarget 2017, 8, 68899–68915. [Google Scholar] [CrossRef] [PubMed]
- Jelonek, K.; Widłak, P. Metabolome-based biomarkers: Their potential role in the early detection of lung cancer. Contemp. Oncol. (Pozn. Pol.) 2018, 22, 135–140. [Google Scholar] [CrossRef] [PubMed]
- Corona, G.; Rizzolio, F.; Giordano, A.; Toffoli, G. Pharmaco-metabolomics: An emerging “omics” tool for the personalization of anticancer treatments and identification of new valuable therapeutic targets. J. Cell. Physiol. 2012, 227, 2827–2831. [Google Scholar] [CrossRef] [PubMed]
- Pinu, R.F.; Beale, J.D.; Paten, M.A.; Kouremenos, K.; Swarup, S.; Schirra, J.H.; Wishart, D. Systems Biology and Multi-Omics Integration: Viewpoints from the Metabolomics Research Community. Metabolites 2019, 9, 76. [Google Scholar] [CrossRef] [PubMed]
- Broadhurst, D.; Goodacre, R.; Reinke, S.N.; Kuligowski, J.; Wilson, I.D.; Lewis, M.R.; Dunn, W.B. Guidelines and considerations for the use of system suitability and quality control samples in mass spectrometry assays applied in untargeted clinical metabolomic studies. Metabolomics 2018, 14, 72. [Google Scholar] [CrossRef]
- Johnson, C.H.; Ivanisevic, J.; Siuzdak, G. Metabolomics: Beyond biomarkers and towards mechanisms. Nat. Rev. Mol. Cell Biol. 2016, 17, 451–459. [Google Scholar] [CrossRef] [PubMed]
- Diamandis, E.P. Cancer biomarkers: Can we turn recent failures into success? J. Natl. Cancer Inst. 2010, 102, 1462–1467. [Google Scholar] [CrossRef]
- Cobb, J.; Gall, W.; Adam, K.-P.; Nakhle, P.; Button, E.; Hathorn, J.; Lawton, K.; Milburn, M.; Perichon, R.; Mitchell, M.; et al. A novel fasting blood test for insulin resistance and prediabetes. J. Diabetes Sci. Technol. 2013, 7, 100–110. [Google Scholar] [CrossRef]
- Kussmann, M.; Kaput, J. Translational genomics. Appl. Transl. Genom. 2014, 3, 43–47. [Google Scholar] [CrossRef] [Green Version]
- Venter, J.C.; Adams, M.D.; Myers, E.W.; Li, P.W.; Mural, R.J.; Sutton, G.G.; Smith, H.O.; Yandell, M.; Evans, C.A.; Holt, R.A.; et al. The Sequence of the Human Genome. Science 2001, 291, 1304–1351. [Google Scholar] [CrossRef] [Green Version]
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; FitzHugh, W.; et al. Initial sequencing and analysis of the human genome. Nature 2001, 409, 860–921. [Google Scholar] [PubMed] [Green Version]
- Park, S.T.; Kim, J. Trends in Next-Generation Sequencing and a New Era for Whole Genome Sequencing. Int. Neurourol. J. 2016, 20, S76–S83. [Google Scholar] [CrossRef] [PubMed]
- Iwamoto, N.; Shimada, T. Recent advances in mass spectrometry-based approaches for proteomics and biologics: Great contribution for developing therapeutic antibodies. Pharmacol. Ther. 2018, 185, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Casamassimi, A.; Federico, A.; Rienzo, M.; Esposito, S.; Ciccodicola, A. Transcriptome Profiling in Human Diseases: New Advances and Perspectives. Int. J. Mol. Sci. 2017, 18, 1652. [Google Scholar] [CrossRef] [PubMed]
- Bossuyt, P.M. Where Are All the New Omics-Based Tests? Clin. Chem. 2014, 60, 1256–1257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diamandis, E.P. The hundred person wellness project and Google’s baseline study: Medical revolution or unnecessary and potentially harmful over-testing? BMC Med. 2015, 13, 5. [Google Scholar] [CrossRef] [PubMed]
- Hayes, D.F. From genome to bedside: Are we lost in translation? Breast 2013, 22, S22–S26. [Google Scholar] [CrossRef] [PubMed]
- Schully, S.D.; Khoury, M.J. What is translational genomics? An expanded research agenda for improving individual and population health. Appl. Transl. Genom. 2014, 3, 82–83. [Google Scholar] [CrossRef] [Green Version]
- McShane, L.M.; Cavenagh, M.M.; Lively, T.G.; Eberhard, D.A.; Bigbee, W.L.; Williams, P.M.; Mesirov, J.P.; Polley, M.-Y.C.; Kim, K.Y.; Tricoli, J.V.; et al. Criteria for the use of omics-based predictors in clinical trials. Nature 2013, 502, 317–320. [Google Scholar] [CrossRef] [Green Version]
- McShane, L.M.; Cavenagh, M.M.; Lively, T.G.; Eberhard, D.A.; Bigbee, W.L.; Williams, P.M.; Mesirov, J.P.; Polley, M.-Y.C.; Kim, K.Y.; Tricoli, J.V.; et al. Criteria for the use of omics-based predictors in clinical trials: Explanation and elaboration. BMC Med. 2013, 11, 220. [Google Scholar] [CrossRef]
- Institute for Systems Biology: 100K Wellness Project. Available online: http://research.systemsbiology.net/100k (accessed on 21 April 2019).
- Institute of Medicine. Evolution of Translational Omics: Lessons Learned and the Path Forward; The National Academies Press: Washington, DC, USA, 2012; p. 354. [Google Scholar] [CrossRef]
- Grattapaglia, D.; Silva-Junior, O.B.; Resende, R.T.; Cappa, E.P.; Müller, B.S.F.; Tan, B.; Isik, F.; Ratcliffe, B.; El-Kassaby, Y.A. Quantitative Genetics and Genomics Converge to Accelerate Forest Tree Breeding. Front. Plant Sci. 2018, 9, 1693. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, A. A Strategy for Identifying Quantitative Trait Genes Using Gene Expression Analysis and Causal Analysis. Genes 2017, 8, 347. [Google Scholar] [CrossRef] [PubMed]
- Phillips, K.A.; Deverka, P.A.; Hooker, G.W.; Douglas, M.P. Genetic Test Availability And Spending: Where Are We Now? Where Are We Going? Health Aff. (Proj. Hope) 2018, 37, 710–716. [Google Scholar] [CrossRef] [Green Version]
- Splinter, K.; Adams, D.R.; Bacino, C.A.; Bellen, H.J.; Bernstein, J.A.; Cheatle-Jarvela, A.M.; Eng, C.M.; Esteves, C.; Gahl, W.A.; Hamid, R.; et al. Effect of Genetic Diagnosis on Patients with Previously Undiagnosed Disease. N. Engl. J. Med. 2018, 379, 2131–2139. [Google Scholar] [CrossRef] [PubMed]
- Bombard, Y. Translating personalized genomic medicine into clinical practice: Evidence, values, and health policy. Genome 2015, 58, 491–497. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, R.; Pasic, M.; Yousef, G.M. Omics for personalized medicine: Defining the current we swim in. Expert Rev. Mol. Diagn. 2016, 16, 719–722. [Google Scholar] [CrossRef] [PubMed]
- Joyner, M.J.; Paneth, N. Seven Questions for Personalized MedicineSeven Questions for Personalized MedicineSeven Questions for Personalized Medicine. JAMA 2015, 314, 999–1000. [Google Scholar] [CrossRef] [PubMed]
- Poste, G. Bring on the biomarkers. Nature 2011, 469, 156–157. [Google Scholar] [CrossRef] [PubMed]
- Lowe, R.; Shirley, N.; Bleackley, M.; Dolan, S.; Shafee, T. Transcriptomics technologies. PLoS Comput. Biol. 2017, 13, e1005457. [Google Scholar] [CrossRef]
- Enquobahrie, D.A.; Rice, K.; Williams, O.D.; Williams, M.A.; Gross, M.D.; Lewis, C.E.; Schwartz, S.M.; Siscovick, D.S. IL1B genetic variation and plasma C-reactive protein level among young adults: The CARDIA study. Atherosclerosis 2009, 202, 513–520. [Google Scholar] [CrossRef] [Green Version]
- Mamtani, M.; Matsubara, T.; Shimizu, C.; Furukawa, S.; Akagi, T.; Onouchi, Y.; Hata, A.; Fujino, A.; He, W.; Ahuja, S.K.; et al. Association of CCR2-CCR5 Haplotypes and CCL3L1 Copy Number with Kawasaki Disease, Coronary Artery Lesions, and IVIG Responses in Japanese Children. PLoS ONE 2010, 5, e11458. [Google Scholar] [CrossRef] [PubMed]
- Carter, C.J. Convergence of genes implicated in Alzheimer’s disease on the cerebral cholesterol shuttle: APP, cholesterol, lipoproteins, and atherosclerosis. Neurochem. Int. 2007, 50, 12–38. [Google Scholar] [CrossRef]
- Whitfield, J.B.; Zhu, G.; Madden, P.A.F.; Montgomery, G.W.; Heath, A.C.; Martin, N.G. Biomarker and Genomic Risk Factors for Liver Function Test Abnormality in Hazardous Drinkers. Alcohol. Clin. Exp. Res. 2019, 43, 473–482. [Google Scholar] [CrossRef] [PubMed]
- Dubot, C.; Bernard, V.; Sablin, M.P.; Vacher, S.; Chemlali, W.; Schnitzler, A.; Pierron, G.; Ait Rais, K.; Bessoltane, N.; Jeannot, E.; et al. Comprehensive genomic profiling of head and neck squamous cell carcinoma reveals FGFR1 amplifications and tumour genomic alterations burden as prognostic biomarkers of survival. Eur. J. Cancer 2018, 91, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Gatalica, Z.; Xiu, J.; Swensen, J.; Vranic, S. Comprehensive analysis of cancers of unknown primary for the biomarkers of response to immune checkpoint blockade therapy. Eur. J. Cancer 2018, 94, 179–186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ergören, M.C.; Söyler, G.; Sah, H.; Becer, E. Investigation of potential genomic biomarkers for obesity and personalized medicine. Int. J. Biol. Macromol. 2019, 122, 493–498. [Google Scholar] [CrossRef]
- Li, H.-H.; Chen, R.; Hyduke, D.R.; Williams, A.; Frötschl, R.; Ellinger-Ziegelbauer, H.; O’Lone, R.; Yauk, C.L.; Aubrecht, J.; Fornace, A.J. Development and validation of a high-throughput transcriptomic biomarker to address 21st century genetic toxicology needs. Proc. Natl. Acad. Sci. USA 2017, 114, E10881–E10889. [Google Scholar] [CrossRef] [Green Version]
- Sivula, L.; Vehniäinen, E.-R.; Karjalainen, A.K.; Kukkonen, J.V.K. Toxicity of biomining effluents to Daphnia magna: Acute toxicity and transcriptomic biomarkers. Chemosphere 2018, 210, 304–311. [Google Scholar] [CrossRef] [PubMed]
- Matsuyama, T.; Ishikawa, T.; Takahashi, N.; Yamada, Y.; Yasuno, M.; Kawano, T.; Uetake, H.; Goel, A. Transcriptomic expression profiling identifies ITGBL1, an epithelial to mesenchymal transition (EMT)-associated gene, is a promising recurrence prediction biomarker in colorectal cancer. Mol. Cancer 2019, 18, 19. [Google Scholar] [CrossRef]
- Bhattacharya, S.; Rosenberg, A.F.; Peterson, D.R.; Grzesik, K.; Baran, A.M.; Ashton, J.M.; Gill, S.R.; Corbett, A.M.; Holden-Wiltse, J.; Topham, D.J.; et al. Transcriptomic Biomarkers to Discriminate Bacterial from Nonbacterial Infection in Adults Hospitalized with Respiratory Illness. Sci. Rep. 2017, 7, 6548. [Google Scholar] [CrossRef]
- Nachun, D.; Gao, F.; Isaacs, C.; Strawser, C.; Yang, Z.; Dokuru, D.; Van Berlo, V.; Sears, R.; Farmer, J.; Perlman, S.; et al. Peripheral blood gene expression reveals an inflammatory transcriptomic signature in Friedreich’s ataxia patients. Hum. Mol. Genet. 2018, 27, 2965–2977. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Sagasti, M.T.; Barrutia, O.; Ribas, G.; Garbisu, C.; Becerril, J.M. Early transcriptomic response of Arabidopsis thaliana to polymetallic contamination: Implications for the identification of potential biomarkers of metal exposure. Metallomics 2016, 8, 518–531. [Google Scholar] [CrossRef] [PubMed]
- O’Connell, G.C.; Treadway, M.B.; Tennant, C.S.; Lucke-Wold, N.; Chantler, P.D.; Barr, T.L. Shifts in Leukocyte Counts Drive the Differential Expression of Transcriptional Stroke Biomarkers in Whole Blood. Transl. Stroke Res. 2019, 10, 26–35. [Google Scholar] [CrossRef] [PubMed]
- Mun, S.; Lee, J.; Lim, M.-K.; Lee, Y.-R.; Ihm, C.; Lee, S.H.; Kang, H.-G. Development of a Novel Diagnostic Biomarker Set for Rheumatoid Arthritis Using a Proteomics Approach. BioMed Res. Int. 2018, 2018, 7490723. [Google Scholar] [CrossRef] [PubMed]
- Shan, J.; Sun, Z.; Yang, J.; Xu, J.; Shi, W.; Wu, Y.; Fan, Y.; Li, H. Discovery and preclinical validation of proteomic biomarkers in saliva for early detection of oral squamous cell carcinomas. Oral Dis. 2019, 25, 97–107. [Google Scholar] [CrossRef] [PubMed]
- Landi, C.; Bargagli, E.; Carleo, A.; Refini, R.M.; Bennett, D.; Bianchi, L.; Cillis, G.; Prasse, A.; Bini, L.; Rottoli, P. Bronchoalveolar lavage proteomic analysis in pulmonary fibrosis associated with systemic sclerosis: S100A6 and 14-3-3ε as potential biomarkers. Rheumatology 2018, 58, 165–178. [Google Scholar]
- Zhan, S.H.; Li, J.M.; Wang, T.X.; Ge, W. Quantitative Proteomics Analysis of Sporadic Medullary Thyroid Cancer Reveals FN1 as a Potential Novel Candidate Prognostic Biomarker. Oncologist 2018, 23, 1415–1425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guzman, Y.A.; Sakellari, D.; Papadimitriou, K.; Floudas, C.A. High-throughput proteomic analysis of candidate biomarker changes in gingival crevicular fluid after treatment of chronic periodontitis. J. Periodontal Res. 2018, 53, 853–860. [Google Scholar] [CrossRef]
- Yang, Y.; Wei, J.; Huang, X.; Wu, M.; Lv, Z.; Tong, P.; Chang, R. iTRAQ-Based Proteomics of Chronic Renal Failure Rats after FuShengong Decoction Treatment Reveals Haptoglobin and Alpha-1-Antitrypsin as Potential Biomarkers. Evid.-Based Complementary Altern. Med. eCAM 2017, 2017, 1480514. [Google Scholar] [CrossRef]
- Turnier, J.L.; Brunner, H.I.; Bennett, M.; Aleed, A.; Gulati, G.; Haffey, W.D.; Thornton, S.; Wagner, M.; Devarajan, P.; Witte, D.; et al. Discovery of SERPINA3 as a candidate urinary biomarker of lupus nephritis activity. Rheumatology 2018, 58, 321–330. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; McCormick, T.S.; Ward, N.L.; Cooper, K.D.; Conic, R.; Xu, R. Combining mechanism-based prediction with patient-based profiling for psoriasis metabolomics biomarker discovery. AMIA Annu. Symp. Proc. 2018, 2017, 1734–1743. [Google Scholar] [PubMed]
- Ban, G.Y.; Cho, K.; Kim, S.H.; Yoon, M.K.; Kim, J.H.; Lee, H.Y.; Shin, Y.S.; Ye, Y.M.; Cho, J.Y.; Park, H.S. Metabolomic analysis identifies potential diagnostic biomarkers for aspirin-exacerbated respiratory disease. Clin. Exp. Allergy 2017, 47, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Gapstur, S.M.; Carter, B.D.; Hartman, T.J.; Stevens, V.L.; Gaudet, M.M.; McCullough, M.L. Untargeted Metabolomics Identifies Novel Potential Biomarkers of Habitual Food Intake in a Cross-Sectional Study of Postmenopausal Women. J. Nutr. 2018, 148, 932–943. [Google Scholar] [CrossRef] [PubMed]
- Potratz, S.; Tarnow, P.; Jungnickel, H.; Baumann, S.; von Bergen, M.; Tralau, T.; Luch, A. Combination of Metabolomics with Cellular Assays Reveals New Biomarkers and Mechanistic Insights on Xenoestrogenic Exposures in MCF-7 Cells. Chem. Res. Toxicol. 2017, 30, 883–892. [Google Scholar] [CrossRef] [PubMed]
- Melvin, S.D.; Lanctôt, C.M.; Doriean, N.J.C.; Carroll, A.R.; Bennett, W.W. Untargeted NMR-based metabolomics for field-scale monitoring: Temporal reproducibility and biomarker discovery in mosquitofish (Gambusia holbrooki) from a metal(loid)-contaminated wetland. Environ. Pollut. 2018, 243, 1096–1105. [Google Scholar] [CrossRef] [PubMed]
- Qiu, S.; Yang, W.-Z.; Yao, C.-L.; Qiu, Z.-D.; Shi, X.-J.; Zhang, J.-X.; Hou, J.-J.; Wang, Q.-R.; Wu, W.-Y.; Guo, D.-A. Nontargeted metabolomic analysis and “commercial-homophyletic” comparison-induced biomarkers verification for the systematic chemical differentiation of five different parts of Panax ginseng. J. Chromatogr. A 2016, 1453, 78–87. [Google Scholar] [CrossRef]
- Godoy-Vitorino, F.; Ortiz-Morales, G.; Romaguera, J.; Sanchez, M.M.; Martinez-Ferrer, M.; Chorna, N. Discriminating high-risk cervical Human Papilloma Virus infections with urinary biomarkers via non-targeted GC-MS-based metabolomics. PLoS ONE 2018, 13, e0209936. [Google Scholar] [CrossRef]
- Schrimpe-Rutledge, A.C.; Codreanu, S.G.; Sherrod, S.D.; McLean, J.A. Untargeted Metabolomics Strategies—Challenges and Emerging Directions. J. Am. Soc. Mass Spectrom. 2016, 27, 1897–1905. [Google Scholar] [CrossRef]
- Zhou, J.; Yin, Y. Strategies for large-scale targeted metabolomics quantification by liquid chromatography-mass spectrometry. Analyst 2016, 141, 6362–6373. [Google Scholar] [CrossRef]
- Lu, W.; Bennett, B.D.; Rabinowitz, J.D. Analytical strategies for LC-MS-based targeted metabolomics. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2008, 871, 236–242. [Google Scholar] [CrossRef]
- Goldansaz, S.A.; Guo, A.C.; Sajed, T.; Steele, M.A.; Plastow, G.S.; Wishart, D.S. Livestock metabolomics and the livestock metabolome: A systematic review. PLoS ONE 2017, 12, e0177675. [Google Scholar] [CrossRef] [PubMed]
- Alonso, A.; Marsal, S.; Julià, A. Analytical Methods in Untargeted Metabolomics: State of the Art in 2015. Front. Bioeng. Biotechnol. 2015, 3, 23. [Google Scholar] [CrossRef] [PubMed]
- Chetwynd, A.J.; Dunn, W.B.; Rodriguez-Blanco, G. Collection and Preparation of Clinical Samples for Metabolomics. In Metabolomics: From Fundamentals to Clinical Applications; Sussulini, A., Ed.; Springer International Publishing: Cham, Switzerland, 2017; pp. 19–44. [Google Scholar]
- Wishart, D.S. Advances in metabolite identification. Bioanalysis 2011, 3, 1769–1782. [Google Scholar] [CrossRef] [PubMed]
- Fiehn, O.; Robertson, D.; Griffin, J.; van der Werf, M.; Nikolau, B.; Morrison, N.; Sumner, L.W.; Goodacre, R.; Hardy, N.W.; Taylor, C.; et al. The metabolomics standards initiative (MSI). Metabolomics 2007, 3, 175–178. [Google Scholar] [CrossRef]
- Pinu, F.R.; Villas-Boas, S.G.; Aggio, R. Analysis of Intracellular Metabolites from Microorganisms: Quenching and Extraction Protocols. Metabolites 2017, 7, 53. [Google Scholar] [CrossRef] [PubMed]
- Villas-Bôas, S.G.; Mas, S.; Åkesson, M.; Smedsgaard, J.; Nielsen, J. Mass spectrometry in metabolome analysis. Mass Spectrom. Rev. 2005, 24, 613–646. [Google Scholar] [CrossRef] [PubMed]
- Pinu, F.R.; Villas-Boas, S.G. Extracellular Microbial Metabolomics: The State of the Art. Metabolites 2017, 7, 43. [Google Scholar] [CrossRef] [PubMed]
- Yurgita, R.V.; Elena, N.L.; Igor, V.U.; Svetlana, D.K.; Antonina, V.S. Metabolomics in Vitamin Status Assessment. Curr. Pharm. Des. 2018, 24, 3028–3033. [Google Scholar]
- Bernini, P.; Bertini, I.; Luchinat, C.; Nincheri, P.; Staderini, S.; Turano, P. Standard operating procedures for pre-analytical handling of blood and urine for metabolomic studies and biobanks. J. Biomol. NMR 2011, 49, 231–243. [Google Scholar] [CrossRef] [PubMed]
- Lei, Z.T.; Huhman, D.V.; Sumner, L.W. Mass Spectrometry Strategies in Metabolomics. J. Biol. Chem. 2011, 286, 25435–25442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sinclair, I.; Bachman, M.; Addison, D.; Rohman, M.; Murray, D.C.; Davies, G.; Mouchet, E.; Tonge, M.E.; Stearns, R.G.; Ghislain, L.; et al. Acoustic Mist Ionization Platform for Direct and Contactless Ultrahigh-Throughput Mass Spectrometry Analysis of Liquid Samples. Anal. Chem. 2019, 91, 3790–3794. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pinu, F.R.; Edwards, P.J.B.; Jouanneau, S.; Kilmartin, P.A.; Gardner, R.C.; Villas-Boas, S.G. Sauvignon blanc metabolomics: Grape juice metabolites affecting the development of varietal thiols and other aroma compounds in wines. Metabolomics 2014, 10, 556–573. [Google Scholar] [CrossRef]
- Zhang, S.; Liu, L.; Steffen, D.; Ye, T.; Raftery, D. Metabolic profiling of gender: Headspace-SPME/GC–MS and 1H NMR analysis of urine. Metabolomics 2012, 8, 323–334. [Google Scholar] [CrossRef]
- Chan, E.C.Y.; Koh, P.K.; Mal, M.; Cheah, P.Y.; Eu, K.W.; Backshall, A.; Cavill, R.; Nicholson, J.K.; Keun, H.C. Metabolic Profiling of Human Colorectal Cancer Using High-Resolution Magic Angle Spinning Nuclear Magnetic Resonance (HR-MAS NMR) Spectroscopy and Gas Chromatography Mass Spectrometry (GC/MS). J. Proteome Res. 2009, 8, 352–361. [Google Scholar] [CrossRef] [PubMed]
- Durand, S.; Sancelme, M.; Besse-Hoggan, P.; Combourieu, B. Biodegradation pathway of mesotrione: Complementarities of NMR, LC–NMR and LC–MS for qualitative and quantitative metabolic profiling. Chemosphere 2010, 81, 372–380. [Google Scholar] [CrossRef] [PubMed]
- Jung, Y.; Ahn, Y.G.; Kim, H.K.; Moon, B.C.; Lee, A.Y.; Ryu, D.H.; Hwang, G.-S. Characterization of dandelion species using 1H NMR- and GC-MS-based metabolite profiling. Analyst 2011, 136, 4222–4231. [Google Scholar] [CrossRef]
- Kim, E.J.; Kwon, J.; Park, S.H.; Park, C.; Seo, Y.-B.; Shin, H.-K.; Kim, H.K.; Lee, K.-S.; Choi, S.-Y.; Ryu, D.H.; et al. Metabolite Profiling of Angelica gigas from Different Geographical Origins Using 1H NMR and UPLC-MS Analyses. J. Agric. Food Chem. 2011, 59, 8806–8815. [Google Scholar] [CrossRef]
- Yang, Z. Online hyphenated liquid chromatography–nuclear magnetic resonance spectroscopy–mass spectrometry for drug metabolite and nature product analysis. J. Pharm. Biomed. Anal. 2006, 40, 516–527. [Google Scholar] [CrossRef]
- Dai, D.; He, J.; Sun, R.; Zhang, R.; Aisa, H.A.; Abliz, Z. Nuclear magnetic resonance and liquid chromatography–mass spectrometry combined with an incompleted separation strategy for identifying the natural products in crude extract. Anal. Chim. Acta 2009, 632, 221–228. [Google Scholar] [CrossRef]
- Stülten, D.; Lamshöft, M.; Zühlke, S.; Spiteller, M. Isolation and characterization of a new human urinary metabolite of diclofenac applying LC–NMR–MS and high-resolution mass analyses. J. Pharm. Biomed. Anal. 2008, 47, 371–376. [Google Scholar] [CrossRef]
- Stoll, D.R.; Carr, P.W. Two-Dimensional Liquid Chromatography: A State of the Art Tutorial. Anal. Chem. 2017, 89, 519–531. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, Z.; Cooks, R.G. Miniature Mass Spectrometers. Annu. Rev. Anal. Chem. 2009, 2, 187–214. [Google Scholar] [CrossRef] [PubMed]
- Snyder, D.T.; Pulliam, C.J.; Ouyang, Z.; Cooks, R.G. Miniature and Fieldable Mass Spectrometers: Recent Advances. Anal. Chem. 2016, 88, 2–29. [Google Scholar] [CrossRef] [PubMed]
- Lu, R.; Zhou, X.; Yin, Q.; Hu, J.; Ni, Z. Miniature nuclear magnetic resonance spectrometer using a partially enclosed permanent magnet. Instrum. Sci. Technol. 2017, 45, 324–337. [Google Scholar] [CrossRef]
- Zhou, X.; Liu, J.; Cooks, R.G.; Ouyang, Z. Development of miniature mass spectrometry systems for bioanalysis outside the conventional laboratories. Bioanalysis 2014, 6, 1497–1508. [Google Scholar] [CrossRef] [Green Version]
- Pinu, F.R.; Tumanov, S.; Grose, C.; Raw, V.; Albright, A.; Stuart, L.; Villas-Boas, S.G.; Martin, D.; Harker, R.; Greven, M. Juice Index: An integrated Sauvignon blanc grape and wine metabolomics database shows mainly seasonal differences. Metabolomics 2019, 15, 3. [Google Scholar] [CrossRef] [PubMed]
- Smart, K.F.; Aggio, R.B.M.; Van Houtte, J.R.; Villas-Bôas, S.G. Analytical platform for metabolome analysis of microbial cells using methyl chloroformate derivatization followed by gas chromatography–mass spectrometry. Nat. Protoc. 2010, 5, 1709–1729. [Google Scholar] [CrossRef] [PubMed]
- Guijas, C.; Montenegro-Burke, J.R.; Domingo-Almenara, X.; Palermo, A.; Warth, B.; Hermann, G.; Koellensperger, G.; Huan, T.; Uritboonthai, W.; Aisporna, A.E.; et al. METLIN: A Technology Platform for Identifying Knowns and Unknowns. Anal. Chem. 2018, 90, 3156–3164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wishart, D.S. Computational strategies for metabolite identification in metabolomics. Bioanalysis 2009, 1, 1579–1596. [Google Scholar] [CrossRef]
- Kvitvang, H.F.N.; Andreassen, T.; Adam, T.; Villas-Bôas, S.G.; Bruheim, P. Highly Sensitive GC/MS/MS Method for Quantitation of Amino and Nonamino Organic Acids. Anal. Chem. 2011, 83, 2705–2711. [Google Scholar] [CrossRef] [PubMed]
- Vielhauer, O.; Zakhartsev, M.; Horn, T.; Takors, R.; Reuss, M. Simplified absolute metabolite quantification by gas chromatography–isotope dilution mass spectrometry on the basis of commercially available source material. J. Chromatogr. B 2011, 879, 3859–3870. [Google Scholar] [CrossRef] [PubMed]
- Hu, K.; Westler, W.M.; Markley, J.L. Simultaneous Quantification and Identification of Individual Chemicals in Metabolite Mixtures by Two-Dimensional Extrapolated Time-Zero 1H−13C HSQC (HSQC0). J. Am. Chem. Soc. 2011, 133, 1662–1665. [Google Scholar] [CrossRef] [PubMed]
- Bennett, B.D.; Yuan, J.; Kimball, E.H.; Rabinowitz, J.D. Absolute quantitation of intracellular metabolite concentrations by an isotope ratio-based approach. Nat. Protoc. 2008, 3, 1299–1311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lien, S.K.; Kvitvang, H.F.N.; Bruheim, P. Utilization of a deuterated derivatization agent to synthesize internal standards for gas chromatography–tandem mass spectrometry quantification of silylated metabolites. J. Chromatogr. A 2012, 1247, 118–124. [Google Scholar] [CrossRef] [PubMed]
- Martineau, E.; Tea, I.; Akoka, S.; Giraudeau, P. Absolute quantification of metabolites in breast cancer cell extracts by quantitative 2D 1H INADEQUATE NMR. NMR Biomed. 2012, 25, 985–992. [Google Scholar] [CrossRef] [PubMed]
- Tumanov, S.; Zubenko, Y.; Obolonkin, V.; Greenwood, D.R.; Shmanai, V.; Villas-Bôas, S.G. Calibration curve-free GC–MS method for quantitation of amino and non-amino organic acids in biological samples. Metabolomics 2016, 12, 64. [Google Scholar] [CrossRef]
- Torii, Y.; Kawano, Y.; Sato, H.; Sasaki, K.; Fujimori, T.; Kawada, J.-I.; Takikawa, O.; Lim, C.K.; Guillemin, G.J.; Ohashi, Y.; et al. Quantitative metabolome profiling reveals the involvement of the kynurenine pathway in influenza-associated encephalopathy. Metabolomics 2016, 12, 84. [Google Scholar] [CrossRef]
- Sun, H.-Z.; Wang, D.-M.; Wang, B.; Wang, J.-K.; Liu, H.-Y.; Guan, L.L.; Liu, J.-X. Metabolomics of Four Biofluids from Dairy Cows: Potential Biomarkers for Milk Production and Quality. J. Proteome Res. 2015, 14, 1287–1298. [Google Scholar] [CrossRef] [PubMed]
- Ghazi, N.; Arjmand, M.; Akbari, Z.; Mellati, A.O.; Saheb-Kashaf, H.; Zamani, Z. (1)H NMR- based metabolomics approaches as non- invasive tools for diagnosis of endometriosis. Int. J. Reprod. Biomed. (YazdIran) 2016, 14, 1–8. [Google Scholar] [CrossRef]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.M.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metab. Off. J. Metab. Soc. 2007, 3, 211–221. [Google Scholar]
- Fernie, A.R.; Aharoni, A.; Willmitzer, L.; Stitt, M.; Tohge, T.; Kopka, J.; Carroll, A.J.; Saito, K.; Fraser, P.D.; DeLuca, V. Recommendations for Reporting Metabolite Data. Plant Cell 2011, 23, 2477–2482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sansone, S.A.; Fan, T.; Goodacre, R.; Griffin, J.L.; Hardy, N.W.; Kaddurah-Daouk, R.; Kristal, B.S.; Lindon, J.; Mendes, P.; Morrison, N.; et al. The metabolomics standards initiative. Nat. Biotechnol. 2007, 25, 846–848. [Google Scholar] [PubMed]
- Xia, J.; Broadhurst, D.I.; Wilson, M.; Wishart, D.S. Translational biomarker discovery in clinical metabolomics: An introductory tutorial. Metab. Off. J. Metab. Soc. 2013, 9, 280–299. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Mandal, R.; Stanislaus, A.; Ramirez-Gaona, M. Cancer Metabolomics and the Human Metabolome Database. Metabolites 2016, 6, 10. [Google Scholar] [CrossRef] [PubMed]
- Dias, D.A.; Hill, C.B.; Jayasinghe, N.S.; Atieno, J.; Sutton, T.; Roessner, U. Quantitative profiling of polar primary metabolites of two chickpea cultivars with contrasting responses to salinity. J. Chromatogr. B 2015, 1000, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zukunft, S.; Prehn, C.; Röhring, C.; Möller, G.; Hrabě de Angelis, M.; Adamski, J.; Tokarz, J. High-throughput extraction and quantification method for targeted metabolomics in murine tissues. Metab. Off. J. Metab. Soc. 2018, 14, 18. [Google Scholar] [CrossRef] [PubMed]
- Jedlicka, L.D.; Silva, J.D.; Balbino, A.M.; Neto, G.B.; Furtado, D.Z.; Silva, H.D.; Cavalcanti, F.D.; Heijden, K.M.; Penatti, C.A.; Bechara, E.J.; et al. Effects of Diacetyl Flavoring Exposure in Mice Metabolism. BioMed Res. Int. 2018, 2018, 9875319. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.; Liang, G.; Zilong, G.; Wensheng, W.; Hongyan, Z.; Xianqing, L.; Sibin, Y.; Lizhong, X.; Jie, L. A Novel Integrated Method for Large-Scale Detection, Identification, and Quantification of Widely Targeted Metabolites: Application in the Study of Rice Metabolomics. Mol. Plant 2013, 6, 1769–1780. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Zhou, L.; Lei, H.; Hao, F.; Liu, X.; Wang, Y.; Tang, H. Simultaneous Quantification of Amino Metabolites in Multiple Metabolic Pathways Using Ultra-High Performance Liquid Chromatography with Tandem-mass Spectrometry. Sci. Rep. 2017, 7, 1423. [Google Scholar] [CrossRef]
- Zhou, J.; Liu, C.; Si, D.; Jia, B.; Zhong, L.; Yin, Y. Workflow development for targeted lipidomic quantification using parallel reaction monitoring on a quadrupole-time of flight mass spectrometry. Anal. Chim. Acta 2017, 972, 62–72. [Google Scholar] [CrossRef]
- Kumar, D.; Thakur, K.; Sharma, S.; Kumar, S. NMR for metabolomics studies of Crataegus rhipidophylla Gand. Anal. Bioanal. Chem. 2019, 411, 2149–2159. [Google Scholar] [CrossRef] [PubMed]
- Gogiashvili, M.; Horsch, S.; Marchan, R.; Gianmoena, K.; Cadenas, C.; Tanner, B.; Naumann, S.; Ersova, D.; Lippek, F.; Rahnenführer, J.; et al. Impact of intratumoral heterogeneity of breast cancer tissue on quantitative metabolomics using high-resolution magic angle spinning 1H NMR spectroscopy. NMR Biomed. 2018, 31, e3862. [Google Scholar] [CrossRef] [PubMed]
- Snyder, N.W.; Mesaros, C.; Blair, I.A. Translational metabolomics in cancer research. Biomark. Med. 2015, 9, 821–834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koczula, K.M.; Gallotta, A. Lateral flow assays. Essays Biochem. 2016, 60, 111–120. [Google Scholar] [CrossRef]
- Deng, L.; Chang, D.; Foshaug, R.R.; Eisner, R.; Tso, K.V.; Wishart, S.D.; Fedorak, N.R. Development and Validation of a High-Throughput Mass Spectrometry Based Urine Metabolomic Test for the Detection of Colonic Adenomatous Polyps. Metabolites 2017, 7, 32. [Google Scholar] [CrossRef] [PubMed]
- Abelson, K.S.P.; Kalliokoski, O.; Teilmann, A.C.; Hau, J. Applicability of Commercially Available ELISA Kits for the Quantification of Faecal Immunoreactive Corticosterone Metabolites in Mice. In Vivo 2016, 30, 739–744. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, S.; Putalun, W.; Vimolmangkang, S.; Phoolcharoen, W.; Shoyama, Y.; Tanaka, H.; Morimoto, S. Enzyme-linked immunosorbent assay for the quantitative/qualitative analysis of plant secondary metabolites. J. Nat. Med. 2018, 72, 32–42. [Google Scholar] [CrossRef] [PubMed]
- Mohr, A.L.A.; Ofsa, B.; Keil, A.M.; Simon, J.R.; McMullin, M.; Logan, B.K. Enzyme-Linked Immunosorbent Assay (ELISA) for the Detection of Use of the Synthetic Cannabinoid Agonists UR-144 and XLR-11 in Human Urine. J. Anal. Toxicol. 2014, 38, 427–431. [Google Scholar] [CrossRef] [Green Version]
- Lu, W.; Su, X.; Klein, M.S.; Lewis, I.A.; Fiehn, O.; Rabinowitz, J.D. Metabolite Measurement: Pitfalls to Avoid and Practices to Follow. Annu. Rev. Biochem. 2017, 86, 277–304. [Google Scholar] [CrossRef]
- Zhang, S.Y.; Garcia-D’Angeli, A.; Brennan, J.P.; Huo, Q. Predicting detection limits of enzyme-linked immunosorbent assay (ELISA) and bioanalytical techniques in general. Analyst 2014, 139, 439–445. [Google Scholar] [CrossRef]
- Daemen, A.; Peterson, D.; Sahu, N.; McCord, R.; Du, X.; Liu, B.; Kowanetz, K.; Hong, R.; Moffat, J.; Gao, M.; et al. Metabolite profiling stratifies pancreatic ductal adenocarcinomas into subtypes with distinct sensitivities to metabolic inhibitors. Proc. Natl. Acad. Sci. USA 2015, 112, E4410–E4417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pepe, M.S.; Feng, Z.; Janes, H.; Bossuyt, P.M.; Potter, J.D. Pivotal evaluation of the accuracy of a biomarker used for classification or prediction: Standards for study design. J. Natl. Cancer Inst. 2008, 100, 1432–1438. [Google Scholar] [CrossRef] [PubMed]
- Drucker, E.; Krapfenbauer, K. Pitfalls and limitations in translation from biomarker discovery to clinical utility in predictive and personalised medicine. EPMA J. 2013, 4, 7. [Google Scholar] [CrossRef] [PubMed]
- Sechidis, K.; Papangelou, K.; Metcalfe, P.D.; Svensson, D.; Weatherall, J.; Brown, G. Distinguishing prognostic and predictive biomarkers: An information theoretic approach. Bioinformatics 2018, 34, 3365–3376. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.J.; Larson, M.G.; Vasan, R.S.; Cheng, S.; Rhee, E.P.; McCabe, E.; Lewis, G.D.; Fox, C.S.; Jacques, P.F.; Fernandez, C.; et al. Metabolite profiles and the risk of developing diabetes. Nat. Med. 2011, 17, 448–453. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.J.; Ngo, D.; Psychogios, N.; Dejam, A.; Larson, M.G.; Vasan, R.S.; Ghorbani, A.; O’Sullivan, J.; Cheng, S.; Rhee, E.P.; et al. 2-Aminoadipic acid is a biomarker for diabetes risk. J. Clin. Investig. 2013, 123, 4309–4317. [Google Scholar] [CrossRef] [PubMed]
- Palmer, N.D.; Stevens, R.D.; Antinozzi, P.A.; Anderson, A.; Bergman, R.N.; Wagenknecht, L.E.; Newgard, C.B.; Bowden, D.W. Metabolomic Profile Associated With Insulin Resistance and Conversion to Diabetes in the Insulin Resistance Atherosclerosis Study. J. Clin. Endocrinol. Metab. 2015, 100, E463–E468. [Google Scholar] [CrossRef]
- Baraldi, E.; Carraro, S.; Giordano, G.; Reniero, F.; Perilongo, G.; Zacchello, F. Metabolomics: Moving towards personalized medicine. Ital. J. Pediatrics 2009, 35, 30. [Google Scholar] [CrossRef]
- Chen, R.; Snyder, M. Promise of personalized omics to precision medicine. Wiley Interdiscip. Rev. Syst. Biol. Med. 2013, 5, 73–82. [Google Scholar] [CrossRef]
- Mastrangelo, A.; Armitage, E.G.; Garcia, A.; Barbas, C. Metabolomics as a Tool for Drug Discovery and Personalised Medicine. A Review. Curr. Top. Med. Chem. 2014, 14, 2627–2636. [Google Scholar] [CrossRef]
- Gibney, M.J.; Walsh, M.; Brennan, L.; Roche, H.M.; German, B.; van Ommen, B. Metabolomics in human nutrition: Opportunities and challenges. Am. J. Clin. Nutr. 2005, 82, 497–503. [Google Scholar] [CrossRef] [PubMed]
- Tebani, A.; Bekri, S. Paving the Way to Precision Nutrition Through Metabolomics. Front. Nutr. 2019, 6, 41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Toro-Martín, J.; Arsenault, B.J.; Després, J.-P.; Vohl, M.-C. Precision Nutrition: A Review of Personalized Nutritional Approaches for the Prevention and Management of Metabolic Syndrome. Nutrients 2017, 9, 913. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.P.; Park, Y.; Ziegler, T.R. Nutritional metabolomics: Progress in addressing complexity in diet and health. Annu. Rev. Nutr. 2012, 32, 183–202. [Google Scholar] [CrossRef] [PubMed]
- Tolstikov, V. Metabolomics: Bridging the Gap between Pharmaceutical Development and Population Health. Metabolites 2016, 6, 20. [Google Scholar] [CrossRef] [PubMed]
- Huan, C.; Wei-na, C.; Sunggoan, J.; Sarah, R.; Stuart, M.; Bronwen, M. Metabolic Dysfunction in Alzheimers Disease and Related Neurodegenerative Disorders. Curr. Alzheimer Res. 2012, 9, 5–17. [Google Scholar]
- Mapstone, M.; Cheema, A.K.; Fiandaca, M.S.; Zhong, X.; Mhyre, T.R.; MacArthur, L.H.; Hall, W.J.; Fisher, S.G.; Peterson, D.R.; Haley, J.M.; et al. Plasma phospholipids identify antecedent memory impairment in older adults. Nat. Med. 2014, 20, 415–418. [Google Scholar] [CrossRef] [PubMed]
- Koeth, R.A.; Wang, Z.; Levison, B.S.; Buffa, J.A.; Org, E.; Sheehy, B.T.; Britt, E.B.; Fu, X.; Wu, Y.; Li, L.; et al. Intestinal microbiota metabolism of l-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat. Med. 2013, 19, 576–585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, J.M.; Hazen, S.L. The Gut Microbial Endocrine Organ: Bacterially Derived Signals Driving Cardiometabolic Diseases. Annu. Rev. Med. 2015, 66, 343–359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pusapati, R.V.; Daemen, A.; Wilson, C.; Sandoval, W.; Gao, M.; Haley, B.; Baudy, A.R.; Hatzivassiliou, G.; Evangelista, M.; Settleman, J. mTORC1-Dependent Metabolic Reprogramming Underlies Escape from Glycolysis Addiction in Cancer Cells. Cancer Cell 2016, 29, 548–562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nicholson, J.K.; Connelly, J.; Lindon, J.C.; Holmes, E. Metabonomics: A platform for studying drug toxicity and gene function. Nat. Rev. Drug Discov. 2002, 1, 153–161. [Google Scholar] [CrossRef] [PubMed]
- Son, H.-S.; Hwang, G.-S.; Kim, K.M.; Ahn, H.-J.; Park, W.-M.; Van Den Berg, F.; Hong, Y.-S.; Lee, C.-H. Metabolomic Studies on Geographical Grapes and Their Wines Using 1H NMR Analysis Coupled with Multivariate Statistics. J. Agric. Food Chem. 2009, 57, 1481–1490. [Google Scholar] [CrossRef] [PubMed]
- Pinu, F.R.; Villas-Boas, S.G.; Martin, D. Pre-fermentative supplementation of fatty acids alters the metabolic activity of wine yeasts. Food Res. Int. 2019. [Google Scholar] [CrossRef] [PubMed]
- Son, H.-S.; Hwang, G.-S.; Kim, K.M.; Kim, E.-Y.; van den Berg, F.; Park, W.-M.; Lee, C.-H.; Hong, Y.-S. 1H NMR-Based Metabolomic Approach for Understanding the Fermentation Behaviors of Wine Yeast Strains. Anal. Chem. 2009, 81, 1137–1145. [Google Scholar] [CrossRef] [PubMed]
- Spevacek, A.R.; Benson, K.H.; Bamforth, C.W.; Slupsky, C.M. Beer metabolomics: Molecular details of the brewing process and the differential effects of late and dry hopping on yeast purine metabolism. J. Inst. Brew. 2016, 122, 21–28. [Google Scholar] [CrossRef]
- Yamamoto, S.; Bamba, T.; Sano, A.; Kodama, Y.; Imamura, M.; Obata, A.; Fukusaki, E. Metabolite profiling of soy sauce using gas chromatography with time-of-flight mass spectrometry and analysis of correlation with quantitative descriptive analysis. J. Biosci. Bioeng. 2012, 114, 170–175. [Google Scholar] [CrossRef] [PubMed]
- Abdelnur, P.V.; Caldana, C.; Martins, M.C.M. Metabolomics applied in bioenergy. Chem. Biol. Technol. Agric. 2014, 1, 22. [Google Scholar] [CrossRef] [Green Version]
- Martien, J.I.; Amador-Noguez, D. Recent applications of metabolomics to advance microbial biofuel production. Curr. Opin. Biotechnol. 2017, 43, 118–126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hollinshead, W.; He, L.; Tang, Y.J. Biofuel production: An odyssey from metabolic engineering to fermentation scale-up. Front. Microbiol. 2014, 5, 344. [Google Scholar] [CrossRef]
- Wang, J.; Chen, L.; Tian, X.; Gao, L.; Niu, X.; Shi, M.; Zhang, W. Global Metabolomic and Network analysis of Escherichia coli Responses to Exogenous Biofuels. J. Proteome Res. 2013, 12, 5302–5312. [Google Scholar] [CrossRef]
- Teoh, S.T.; Putri, S.; Mukai, Y.; Bamba, T.; Fukusaki, E. A metabolomics-based strategy for identification of gene targets for phenotype improvement and its application to 1-butanol tolerance in Saccharomyces cerevisiae. Biotechnol. Biofuels 2015, 8, 144. [Google Scholar] [CrossRef]
- Turner, M.F.; Heuberger, A.L.; Kirkwood, J.S.; Collins, C.C.; Wolfrum, E.J.; Broeckling, C.D.; Prenni, J.E.; Jahn, C.E. Non-targeted Metabolomics in Diverse Sorghum Breeding Lines Indicates Primary and Secondary Metabolite Profiles Are Associated with Plant Biomass Accumulation and Photosynthesis. Front. Plant Sci. 2016, 7, 953. [Google Scholar] [CrossRef] [Green Version]
- Liew, F.; Martin, M.E.; Tappel, R.C.; Heijstra, B.D.; Mihalcea, C.; Köpke, M. Gas Fermentation-A Flexible Platform for Commercial Scale Production of Low-Carbon-Fuels and Chemicals from Waste and Renewable Feedstocks. Front. Microbiol. 2016, 7, 694. [Google Scholar] [CrossRef]
- Heijstra, B.D.; Leang, C.; Juminaga, A. Gas fermentation: Cellular engineering possibilities and scale up. Microb. Cell Factories 2017, 16, 60. [Google Scholar] [CrossRef]
- Wan, N.; Sathish, A.; You, L.; Tang, Y.J.; Wen, Z. Deciphering Clostridium metabolism and its responses to bioreactor mass transfer during syngas fermentation. Sci. Rep. 2017, 7, 10090. [Google Scholar] [CrossRef] [Green Version]



| Omics | Candidate Biomarker(s) | Application | Reference |
|---|---|---|---|
| Genomics | IL1B | Obesity | [54] |
| CCL3L1 | Kawasaki Disease, risk of coronary artery lesions and resistance to intravenous immunoglobulin | [55] | |
| GSK3B | Alzheimer's disease | [56] | |
| PNPLA3, TM6SF2, HSD17B13 | Alcoholic liver disease | [57] | |
| TP53, CCND1, CDKN2A, FGFR1 | Head and neck squamous cell carcinoma | [58] | |
| MSI-H, PD-L1, TML-H | Cancer of unknown primary (CUP) | [59] | |
| FTO rs9939609 | Obesity | [60] | |
| Transcriptomics | TGx-DDI | Genotoxicity Screening | [61] |
| Transcriptome factors of enzymes: Monooxygenase, vitellogenin superoxide dismutase, catalase | Metal mixture toxicity | [62] | |
| ITGBL1 | Colorectal cancer | [63] | |
| ICAM1, ITGAL, ITGB2, PECAM1, IGFBP2, IGFBP6, CTSG, MMP2, ACOX3, FADS2, PLA2GA4 | Lower respiratory tract infection | [64] | |
| PI3, CA1, SNCA, FCGBP, GNG10, PROK2, CHPT1, GZMB, CD79A, ALPL | Friedreich’s ataxia | [65] | |
| AOP2, SAUR16, ASN1, DIN2 | Plant early metal exposure | [66] | |
| PLXDC2, STK3, ANTXR2, KIF1B, CD163, CTSZ, PDK4, GRAP, MAL, ID3 | Stroke | [67] | |
| Proteomics | SAA4, gelsolin, vitamin D-binding protein | Rheumatoid arthritis | [68] |
| Solute carrier family 3 member 2, S100 calcium-binding protein A2, interleukin-1 receptor antagonist protein | Oral squamous cell carcinomas | [69] | |
| C3a, APOAI, 14-3-3ε, SPFA2, S100A6 | Systemic sclerosis | [70] | |
| FN1, RPS6KA3 | Sporadic medullary thyroid cancer | [71] | |
| Azurocidin, lysozyme C, myosin-9, alpha-smooth muscle actin | Periodontal disease | [72] | |
| Haptoglobin, alpha-1-antitrypsin | Chronic renal failure and FuShengong Decoction | [73] | |
| SERPINA3 | Lupus nephritis chronicity | [74] | |
| Metabolomics | Linoleic acid, 13(S)-hydroxy-9Z,11E-octadecadienoic acid | Psoriasis | [75] |
| LTE4, LTE4/PGF2a | Aspirin-exacerbated respiratory disease | [76] | |
| Dopamine 3-O-sulfate, dopamine 4-O-sulfate, alliin, N-acetylalliin, S-allylcysteine | Food biomarkers in postmenopausal women | [77] | |
| Proline | Xenoestrogenic exposures in MCF-7 cells | [78] | |
| Aspartate, histidine, myo-inositol, taurine, choline | Metal(loid)-contaminated mosquitofish | [79] | |
| Re, Rg1, Rg2, a flavonoid, Rc, Rf, F1, Ro, vina-R4, acetyl-Rh13/Rh19, floral-I/J | Systematic chemical differentiation of five different parts of Panax ginseng | [80] | |
| 5-Oxoprolinate, Erythronic acid, N-Acetylaspartic acid | Human papilloma virus | [81] |
| Platform | Quantification Method | Number of Metabolites | Targeted/Untargeted | Reference |
|---|---|---|---|---|
| GC-MS | Calibration curve free quantification method using methyl chloroformate derivatisation (MCF) method | 50–100 | Targeted | [121] |
| GC-MS | N,O-bis -(trimethylsilyl)trifluoroacetamide (BSTFA) derivatisation of primary metabolites | 49 | Targeted | [130] |
| GC-MS/MS | MCF derivatisation | 67 | Targeted | [115] |
| LC-MS/MS and FIA-MS/MS,UPLC-MS/MS | AbsoluteIDQ™ p180 Kit (Biocrates) | 188 | Targeted | [131,132] |
| LC–MS | Stepwise multiple ion monitoring-enhanced product ions | 277 | Untargeted | [133] |
| UPLC-MS/MS | Derivatization assisted sensitivity enhancement with 5-aminoisoquinolyl-N-hydroxysuccinimidyl carbamate | 124 | Targeted | [134] |
| QTOF LC-MS | PRM | 222 | Targeted | [135] |
| LC-MRM/PRM-MS | MRM and PRM | 71–387 | Targeted | [83] |
| NMR | Ratio method | 58 | Targeted | [136] |
| NMR | HR MAS | 32 | Targeted | [137] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pinu, F.R.; Goldansaz, S.A.; Jaine, J. Translational Metabolomics: Current Challenges and Future Opportunities. Metabolites 2019, 9, 108. https://doi.org/10.3390/metabo9060108
Pinu FR, Goldansaz SA, Jaine J. Translational Metabolomics: Current Challenges and Future Opportunities. Metabolites. 2019; 9(6):108. https://doi.org/10.3390/metabo9060108
Chicago/Turabian StylePinu, Farhana R., Seyed Ali Goldansaz, and Jacob Jaine. 2019. "Translational Metabolomics: Current Challenges and Future Opportunities" Metabolites 9, no. 6: 108. https://doi.org/10.3390/metabo9060108
APA StylePinu, F. R., Goldansaz, S. A., & Jaine, J. (2019). Translational Metabolomics: Current Challenges and Future Opportunities. Metabolites, 9(6), 108. https://doi.org/10.3390/metabo9060108