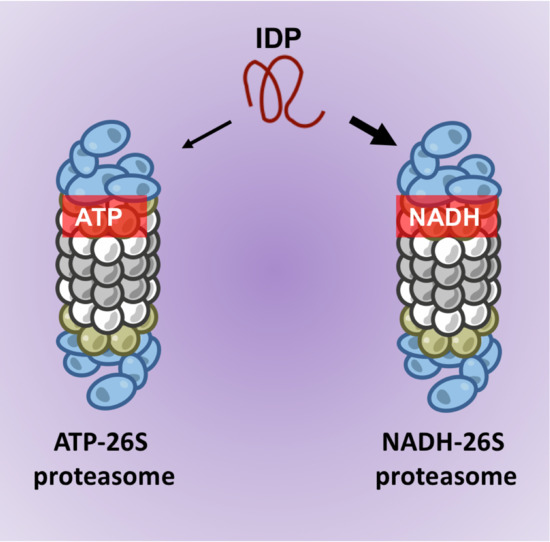
Degradation of Intrinsically Disordered Proteins by the NADH 26S Proteasome
Abstract
:1. Introduction
2. Materials and Methods
2.1. Proteasomal Complex Stability Assay
2.2. Nondenaturing PAGE
2.3. Proteasomal Activity
2.4. 35S In Vitro Translated Proteins and Purification
2.5. In Vitro Degradation Assay
3. Results
3.1. NADH Inhibits ATP-Dependent, 26S Proteasome Degradation of ODC
3.2. ATPγS-26S PC Promotes the Degradation of IDPs
3.3. NADH-26S Proteasomes Can Degrade IDPs
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Hough, R.; Pratt, G.; Rechsteiner, M. Purification of two high molecular weight proteases from rabbit reticulocyte lysate. J. Biol. Chem. 1987, 262, 8303–8313. [Google Scholar] [PubMed]
- Tanaka, K.; Ichihara, A. Involvement of proteasomes (multicatalytic proteinase) in ATP-dependent proteolysis in rat reticulocyte extracts. FEBS Lett. 1988, 236, 159–162. [Google Scholar] [CrossRef] [Green Version]
- Waxman, L.; Fagan, J.M.; Goldberg, A.L. Demonstration of two distinct high molecular weight proteases in rabbit reticulocytes, one of which degrades ubiquitin conjugates. J. Biol. Chem. 1987, 262, 2451–2456. [Google Scholar] [PubMed]
- Glickman, M.H.; Rubin, D.M.; Coux, O.; Wefes, I.; Pfeifer, G.; Cjeka, Z.; Baumeister, W.; A Fried, V.; Finley, D. A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-Signalosome and eIF. Cell 1998, 94, 615–623. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, M.; Finley, D. Regulation of proteasome activity in health and disease. Biochim. Biophys. Acta (BBA)-Bioenerg. 2014, 1843, 13–25. [Google Scholar] [CrossRef] [Green Version]
- Bard, J.A.; Goodall, E.A.; Greene, E.R.; Jonsson, E.; Dong, K.C.; Martin, A. Structure and function of the 26S Proteasome. Annu. Rev. Biochem. 2018, 87, 697–724. [Google Scholar] [CrossRef]
- Collins, G.A.; Goldberg, A.L. The logic of the 26S proteasome. Cell 2017, 169, 792–806. [Google Scholar] [CrossRef] [Green Version]
- Shyu, Y.J.; Liu, H.; Deng, X.; Hu, C.-D. Identification of new fluorescent protein fragments for bimolecular fluorescence complementation analysis under physiological conditions. Biotechniques 2006, 40, 61–66. [Google Scholar] [CrossRef] [Green Version]
- Kleijnen, M.F.; Roelofs, J.; Park, S.; Hathaway, N.A.; Glickman, M.H.; King, R.W.; Finley, D. Stability of the proteasome can be regulated allosterically through engagement of its proteolytic active sites. Nat. Struct. Mol. Biol. 2007, 14, 1180–1188. [Google Scholar] [CrossRef]
- Liu, C.-W.; Li, X.; Thompson, D.; Wooding, K.; Chang, T.-L.; Tang, Z.; Yu, H.; Thomas, P.J.; DeMartino, G.N. ATP binding and ATP hydrolysis play distinct roles in the function of 26s proteasome. Mol. Cell 2006, 24, 39–50. [Google Scholar] [CrossRef] [Green Version]
- Smith, D.M.; Kafri, G.; Cheng, Y.; Ng, D.; Walz, T.; Goldberg, A.L. ATP binding to PAN or the 26S ATPases causes association with the 20S proteasome, gate opening, and translocation of unfolded proteins. Mol. Cell 2005, 20, 687–698. [Google Scholar] [CrossRef] [PubMed]
- Leggett, D.S.; Hanna, J.; Borodovsky, A.; Crosas, B.; Schmidt, M.; Baker, R.T.; Walz, T.; Ploegh, H.; Finley, D. Multiple associated proteins regulate proteasome structure and function. Mol. Cell 2002, 10, 495–507. [Google Scholar] [CrossRef]
- Livnat-Levanon, N.; Kevei, É.; Kleifeld, O.; Krutauz, D.; Segref, A.; Rinaldi, T.; Erpapazoglou, Z.; Cohen, M.; Reis, N.; Hoppe, T.; et al. Reversible 26S proteasome disassembly upon mitochondrial stress. Cell Rep. 2014, 7, 1371–1380. [Google Scholar] [CrossRef] [PubMed]
- Meul, T.; Berschneider, K.; Schmitt, S.; Mayr, C.H.; Mattner, L.F.; Schiller, H.B.; Yazgili, A.S.; Wang, X.; Lukas, C.; Schlesser, C.; et al. Mitochondrial regulation of the 26S proteasome. Cell Rep. 2020, 32, 108059. [Google Scholar] [CrossRef] [PubMed]
- Tsvetkov, P.; Detappe, A.; Cai, K.; Keys, H.R.; Brune, Z.; Ying, W.; Thiru, P.; Reidy, M.; Kugener, G.; Rossen, J.; et al. Author Correction: Mitochondrial metabolism promotes adaptation to proteotoxic stress. Nat. Chem. Biol. 2019, 15, 757. [Google Scholar] [CrossRef] [Green Version]
- Ying, W. NAD+/NADH and NADP+/NADPH in cellular functions and cell death: Regulation and biological consequences. Antioxid. Redox Signal. 2008, 10, 179–206. [Google Scholar] [CrossRef] [Green Version]
- Spinelli, J.B.; Haigis, M.C. The multifaceted contributions of mitochondria to cellular metabolism. Nat. Cell Biol. 2018, 20, 745–754. [Google Scholar] [CrossRef]
- Tsvetkov, P.; Myers, N.; Eliav, R.; Adamovich, Y.; Hagai, T.; Adler, J.; Navon, A.; Shaul, Y. NADH binds and stabilizes the 26S proteasomes independent of ATP. J. Biol. Chem. 2014, 289, 11272–11281. [Google Scholar] [CrossRef] [Green Version]
- Smith, D.M.; Fraga, H.; Reis, C.; Kafri, G.; Goldberg, A.L. ATP binds to proteasomal ATPases in pairs with distinct functional effects, implying an ordered reaction cycle. Cell 2011, 144, 526–538. [Google Scholar] [CrossRef] [Green Version]
- Babu, M.M. The contribution of intrinsically disordered regions to protein function, cellular complexity, and human disease. Biochem. Soc. Trans. 2016, 44, 1185–1200. [Google Scholar] [CrossRef] [Green Version]
- Dunker, A.K.; Obradovic, Z.; Romero, P.; Garner, E.C.; Brown, C.J. Intrinsic protein disorder in complete genomes. Genome Inf. Ser. Workshop Genome Inf. 2000, 11, 161–171. [Google Scholar]
- Uversky, V.N.; Dunker, A.K. Biochemistry: Controlled chaos. Science 2008, 322, 1340–1341. [Google Scholar] [CrossRef] [PubMed]
- Gsponer, J.; Futschik, M.E.; Teichmann, S.A.; Babu, M.M. Tight regulation of unstructured proteins: From transcript synthesis to protein degradation. Science 2008, 322, 1365–1368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsvetkov, P.; Asher, G.; Paz, A.; Reuven, N.; Sussman, J.L.; Silman, I.; Shaul, Y. Operational definition of intrinsically unstructured protein sequences based on susceptibility to the 20S proteasome. Proteins Struct. Funct. Bioinform. 2008, 70, 1357–1366. [Google Scholar] [CrossRef] [PubMed]
- Asher, G.; Reuven, N.; Shaul, Y. 20S proteasomes and protein degradation “by default”. BioEssays 2006, 28, 844–849. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.-W.; Corboy, M.J.; DeMartino, G.N.; Thomas, P.J. Endoproteolytic activity of the proteasome. Science 2003, 299, 408–411. [Google Scholar] [CrossRef] [Green Version]
- Myers, N.; Olender, T.; Savidor, A.; Levin, Y.; Reuven, N.; Shaul, Y. The disordered landscape of the 20S proteasome substrates reveals tight association with phase separated granules. Proteomics 2018, 18, e1800076. [Google Scholar] [CrossRef]
- Tsvetkov, P.; Myers, N.; Moscovitz, O.; Sharon, M.; Prilusky, J.; Shaul, Y. Thermo-resistant intrinsically disordered proteins are efficient 20S proteasome substrates. Mol. Biosyst. 2012, 8, 368–373. [Google Scholar] [CrossRef]
- Murakami, Y.; Matsufuji, S.; Kameji, T.; Hayashi, S.-I.; Igarashi, K.; Tamura, T.; Tanaka, K.; Ichihara, A. Ornithine decarboxylase is degraded by the 26S proteasome without ubiquitination. Nature 1992, 360, 597–599. [Google Scholar] [CrossRef]
- Kahana, C. Protein degradation, the main hub in the regulation of cellular polyamines. Biochem. J. 2016, 473, 4551–4558. [Google Scholar] [CrossRef]
- Coffino, P. Antizyme, a mediator of ubiquitin-independent proteasomal degradation. Biochimie 2001, 83, 319–323. [Google Scholar] [CrossRef]
- Li, X.; DeMartino, G.N. Variably modulated gating of the 26S proteasome by ATP and polyubiquitin. Biochem. J. 2009, 421, 397–404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ou, L.; Waddell, M.B.; Kriwacki, R.W. Mechanism of cell cycle entry mediated by the intrinsically disordered protein P27Kip1. ACS Chem. Biol. 2012, 7, 678–682. [Google Scholar] [CrossRef] [Green Version]
- Tsytlonok, M.; Hemmen, K.; Hamilton, G.; Kolimi, N.; Felekyan, S.; Seidel, C.A.; Tompa, P.; Sanabria, H. Specific conformational dynamics and expansion underpin a multi-step mechanism for specific binding of p27 with Cdk2/Cyclin A. J. Mol. Biol. 2020, 432, 2998–3017. [Google Scholar] [CrossRef] [PubMed]
- Asher, G.; Tsvetkov, P.; Kahana, C.; Shaul, Y. A mechanism of ubiquitin-independent proteasomal degradation of the tumor suppressors p53 and p73. Genes Dev. 2005, 19, 316–321. [Google Scholar] [CrossRef] [Green Version]
- Henderson, A.; Erales, J.; Hoyt, M.A.; Coffino, P. Dependence of proteasome processing rate on substrate unfolding. J. Biol. Chem. 2011, 286, 17495–17502. [Google Scholar] [CrossRef] [Green Version]
- Sauer, R.T.; Baker, T.A. AAA+ proteases: ATP-Fueled machines of protein destruction. Annu. Rev. Biochem. 2011, 80, 587–612. [Google Scholar] [CrossRef]
- Ataullakhanov, F.I.; Vitvitsky, V.M. What determines the intracellular ATP concentration. Biosci. Rep. 2002, 22, 501–511. [Google Scholar] [CrossRef] [Green Version]
- Chandel, N.S. Evolution of mitochondria as signaling organelles. Cell Metab. 2015, 22, 204–206. [Google Scholar] [CrossRef] [Green Version]
- Adamovich, Y.; Shlomai, A.; Tsvetkov, P.; Umansky, K.B.; Reuven, N.; Estall, J.L.; Spiegelman, B.M.; Shaul, Y. The protein level of PGC-1, a key metabolic regulator, is controlled by NADH-Nqomol. Cell. Biol. 2013, 33, 2603–2613. [Google Scholar] [CrossRef] [Green Version]
- Shaul, Y.; Tsvetkov, P.; Reuven, N. IDPs and protein degradation in the cell. In Instrumental Analysis of Intrinsically Disordered Proteins; Vladimir, N.U., Sonia, L., Eds.; John Wiley & Sons: Hoboken, NJ, USA, 2010; pp. 1–36. [Google Scholar]
- Śledź, P.; Unverdorben, P.; Beck, F.; Pfeifer, G.; Schweitzer, A.; Förster, F.; Baumeister, W. Structure of the 26S proteasome with ATP-S bound provides insights into the mechanism of nucleotide-dependent substrate translocation. Proc. Natl. Acad. Sci. USA 2013, 110, 7264–7269. [Google Scholar] [CrossRef] [PubMed] [Green Version]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsvetkov, P.; Myers, N.; Adler, J.; Shaul, Y. Degradation of Intrinsically Disordered Proteins by the NADH 26S Proteasome. Biomolecules 2020, 10, 1642. https://doi.org/10.3390/biom10121642
Tsvetkov P, Myers N, Adler J, Shaul Y. Degradation of Intrinsically Disordered Proteins by the NADH 26S Proteasome. Biomolecules. 2020; 10(12):1642. https://doi.org/10.3390/biom10121642
Chicago/Turabian StyleTsvetkov, Peter, Nadav Myers, Julia Adler, and Yosef Shaul. 2020. "Degradation of Intrinsically Disordered Proteins by the NADH 26S Proteasome" Biomolecules 10, no. 12: 1642. https://doi.org/10.3390/biom10121642
APA StyleTsvetkov, P., Myers, N., Adler, J., & Shaul, Y. (2020). Degradation of Intrinsically Disordered Proteins by the NADH 26S Proteasome. Biomolecules, 10(12), 1642. https://doi.org/10.3390/biom10121642