Highly Sensitive Detection of Zika Virus Nonstructural Protein 1 in Serum Samples by a Two-Site Nanobody ELISA
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
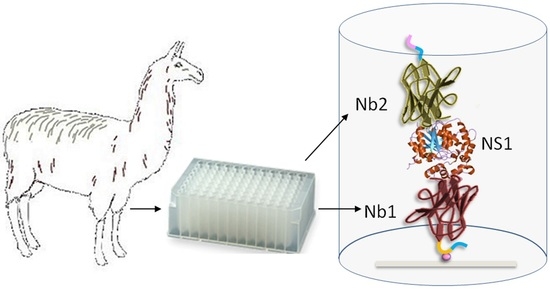
2.2. Llama Immunization and Library Construction
2.3. Purification of Immune Llama Immunoglobulins
2.4. Panning Strategies for the Selection of ZVNS1-Specific Antibodies
2.5. Nanobody Expression
2.6. ELISA Method for Selection of Capturing Nanobodies
2.7. Large-Scale Production of Biotinylated and HA-Tagged Nbs
2.8. Pairwise Selection of Nanobodies
2.9. Nanobody Sandwich ELISA for the Detection of ZVNS1 in Serum Samples
3. Results and Discussion
3.1. To Promote a Broad Representation of the NS1 Epitopes, Different Antigen Immobilization Strategies Were Used to Pan the Nb Library
3.2. Most Capture Nbs Had Negligible Cross-Reactivity with Other Flavivirus NS1 Proteins
3.3. Selection of Best Nanobody Pair for the Detection of ZVNS1
3.4. Development of a Nano-Sandwich ELISA for Quantification of ZVNS1
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Dick, G.W.; Kitchen, S.F.; Haddow, A.J. Zika virus. I. Isolations and serological specificity. Trans. R. Soc. Trop. Med. Hyg. 1952, 46, 509–520. [Google Scholar] [CrossRef]
- Heukelbach, J.; Alencar, C.H.; Kelvin, A.A.; de Oliveira, W.K.; de Goes Cavalcanti, L.P. Zika virus outbreak in Brazil. J. Infect. Dev. Ctries. 2016, 10, 116–120. [Google Scholar] [CrossRef] [PubMed]
- Faria, N.R.; Azevedo, R.; Kraemer, M.U.G.; Souza, R.; Cunha, M.S.; Hill, S.C.; Theze, J.; Bonsall, M.B.; Bowden, T.A.; Rissanen, I.; et al. Zika virus in the Americas: Early epidemiological and genetic findings. Science 2016, 352, 345–349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eppes, C.; Rac, M.; Dunn, J.; Versalovic, J.; Murray, K.O.; Suter, M.A.; Sanz Cortes, M.; Espinoza, J.; Seferovic, M.D.; Lee, W.; et al. Testing for Zika virus infection in pregnancy: Key concepts to deal with an emerging epidemic. Am. J. Obstet. Gynecol. 2017, 216, 209–225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, W.C.; Akey, D.L.; Konwerski, J.R.; Tarrasch, J.T.; Skiniotis, G.; Kuhn, R.J.; Smith, J.L. Extended surface for membrane association in Zika virus NS1 structure. Nat. Struct. Mol. Biol. 2016, 23, 865–867. [Google Scholar] [CrossRef]
- Landry, M.L.; St George, K. Laboratory diagnosis of Zika virus infection. Arch. Pathol. Lab. Med. 2017, 141, 60–67. [Google Scholar] [CrossRef] [Green Version]
- Rossini, G.; Gaibani, P.; Vocale, C.; Cagarelli, R.; Landini, M.P. Comparison of Zika virus (ZIKV) RNA detection in plasma, whole blood and urine—Case series of travel-associated ZIKV infection imported to Italy, 2016. J. Infect. 2017, 75, 242–245. [Google Scholar] [CrossRef] [Green Version]
- Priyamvada, L.; Quicke, K.M.; Hudson, W.H.; Onlamoon, N.; Sewatanon, J.; Edupuganti, S.; Pattanapanyasat, K.; Chokephaibulkit, K.; Mulligan, M.J.; Wilson, P.C.; et al. Human antibody responses after dengue virus infection are highly cross-reactive to Zika virus. Proc. Natl. Acad. Sci. USA 2016, 113, 7852–7857. [Google Scholar] [CrossRef] [Green Version]
- Zaidi, M.B.; Cedillo-Barron, L.; Gonzalez, Y.A.M.E.; Garcia-Cordero, J.; Campos, F.D.; Namorado-Tonix, K.; Perez, F. Serological tests reveal significant cross-reactive human antibody responses to Zika and Dengue viruses in the Mexican population. Acta Trop. 2020, 201, 105201. [Google Scholar] [CrossRef]
- Felix, A.C.; Souza, N.C.S.; Figueiredo, W.M.; Costa, A.A.; Inenami, M.; da Silva, R.M.G.; Levi, J.E.; Pannuti, C.S.; Romano, C.M. Cross reactivity of commercial anti-dengue immunoassays in patients with acute Zika virus infection. J. Med. Virol. 2017, 89, 1477–1479. [Google Scholar] [CrossRef]
- Muller, D.A.; Young, P.R. The flavivirus NS1 protein: Molecular and structural biology, immunology, role in pathogenesis and application as a diagnostic biomarker. Antivir. Res. 2013, 98, 192–208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gutsche, I.; Coulibaly, F.; Voss, J.E.; Salmon, J.; d’Alayer, J.; Ermonval, M.; Larquet, E.; Charneau, P.; Krey, T.; Megret, F.; et al. Secreted dengue virus nonstructural protein NS1 is an atypical barrel-shaped high-density lipoprotein. Proc. Natl. Acad. Sci. USA 2011, 108, 8003–8008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alcon, S.; Talarmin, A.; Debruyne, M.; Falconar, A.; Deubel, V.; Flamand, M. Enzyme-linked immunosorbent assay specific to Dengue virus type 1 nonstructural protein NS1 reveals circulation of the antigen in the blood during the acute phase of disease in patients experiencing primary or secondary infections. J. Clin. Microbiol. 2002, 40, 376–381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allonso, D.; Meneses, M.D.; Fernandes, C.A.; Ferreira, D.F.; Mohana-Borges, R. Assessing positivity and circulating levels of NS1 in samples from a 2012 dengue outbreak in Rio de Janeiro, Brazil. PLoS ONE 2014, 9, e113634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Russo, F.B.; Jungmann, P.; Beltrao-Braga, P.C.B. Zika infection and the development of neurological defects. Cell Microbiol. 2017, 19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andreata-Santos, R.; Pereira, S.S.; Pereira, L.R.; Felix, A.C.; Romano, C.M.; Ferreira, L.C.S. Specificity of NS1-based immunochromatographic tests for dengue virus with regard to the Zika virus protein. Int. J. Infect. Dis. 2020, 95, 276–278. [Google Scholar] [CrossRef]
- Hamers-Casterman, C.; Atarhouch, T.; Muyldermans, S.; Robinson, G.; Hamers, C.; Songa, E.B.; Bendahman, N.; Hamers, R. Naturally occurring antibodies devoid of light chains. Nature 1993, 363, 446–448. [Google Scholar] [CrossRef]
- Gonzalez-Sapienza, G.; Rossotti, M.A.; Tabares-da Rosa, S. Single-domain antibodies as versatile affinity reagents for analytical and diagnostic applications. Front. Immunol. 2017, 8, 977. [Google Scholar] [CrossRef]
- Muyldermans, S. Nanobodies: Natural single-domain antibodies. Annu. Rev. Biochem. 2013, 82, 775–797. [Google Scholar] [CrossRef] [Green Version]
- van der Linden, R.H.; Frenken, L.G.; de Geus, B.; Harmsen, M.M.; Ruuls, R.C.; Stok, W.; de Ron, L.; Wilson, S.; Davis, P.; Verrips, C.T. Comparison of physical chemical properties of llama VHH antibody fragments and mouse monoclonal antibodies. Biochim. Biophys. Acta 1999, 1431, 37–46. [Google Scholar] [CrossRef]
- Rossotti, M.A.; Pirez, M.; Gonzalez-Techera, A.; Cui, Y.; Bever, C.S.; Lee, K.S.; Morisseau, C.; Leizagoyen, C.; Gee, S.; Hammock, B.D.; et al. Method for sorting and pairwise selection of nanobodies for the development of highly sensitive sandwich immunoassays. Anal. Chem. 2015, 87, 11907–11914. [Google Scholar] [CrossRef] [Green Version]
- Delfin-Riela, T.; Rossotti, M.A.; Echaides, C.; Gonzalez-Sapienza, G. A nanobody-based test for highly sensitive detection of hemoglobin in fecal samples. Anal. Bioanal. Chem. 2020, 412, 389–396. [Google Scholar] [CrossRef]
- Tabares-da Rosa, S.; Rossotti, M.; Carleiza, C.; Carrion, F.; Pritsch, O.; Ahn, K.C.; Last, J.A.; Hammock, B.D.; Gonzalez-Sapienza, G. Competitive selection from single domain antibody libraries allows isolation of high-affinity antihapten antibodies that are not favored in the llama immune response. Anal. Chem. 2011, 83, 7213–7220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rossotti, M.; Tabares, S.; Alfaya, L.; Leizagoyen, C.; Moron, G.; Gonzalez-Sapienza, G. Streamlined method for parallel identification of single domain antibodies to membrane receptors on whole cells. Biochim. Biophys. Acta 2015, 1850, 1397–1404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chapman-Smith, A.; Turner, D.L.; Cronan, J.E., Jr.; Morris, T.W.; Wallace, J.C. Expression, biotinylation and purification of a biotin-domain peptide from the biotin carboxy carrier protein of Escherichia coli acetyl-CoA carboxylase. Biochem. J. 1994, 302, 881–887. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harmsen, M.M.; Ruuls, R.C.; Nijman, I.J.; Niewold, T.A.; Frenken, L.G.; de Geus, B. Llama heavy-chain V regions consist of at least four distinct subfamilies revealing novel sequence features. Mol. Immunol. 2000, 37, 579–590. [Google Scholar] [CrossRef]
- Song, H.; Qi, J.; Haywood, J.; Shi, Y.; Gao, G.F. Zika virus NS1 structure reveals diversity of electrostatic surfaces among flaviviruses. Nat. Struct. Mol. Biol. 2016, 23, 456–458. [Google Scholar] [CrossRef] [PubMed]
- Bosch, I.; de Puig, H.; Hiley, M.; Carre-Camps, M.; Perdomo-Celis, F.; Narvaez, C.F.; Salgado, D.M.; Senthoor, D.; O’Grady, M.; Phillips, E.; et al. Rapid antigen tests for dengue virus serotypes and Zika virus in patient serum. Sci. Transl. Med. 2017, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de la Cruz-Hernandez, S.I.; Flores-Aguilar, H.; Gonzalez-Mateos, S.; Lopez-Martinez, I.; Alpuche-Aranda, C.; Ludert, J.E.; del Angel, R.M. Determination of viremia and concentration of circulating nonstructural protein 1 in patients infected with dengue virus in Mexico. Am. J. Trop. Med. Hyg. 2013, 88, 446–454. [Google Scholar] [CrossRef] [Green Version]
- SANTE/11813/2017. Available online: https://ec.europa.eu/food/sites/food/files/plant/docs/pesticides_mrl_guidelines_wrkdoc_2017-11813.pdf (accessed on 24 November 2020).








| Unspiked | Spiked ZVNS1 1.50 ng/mL | Spiked ZVNS1 4.50 ng/mL | |||
|---|---|---|---|---|---|
| Sample | ZVNS1 ng/mL | ZVNS1 ng/mL | % Recovery | ZVNS1 ng/mL | % Recovery |
| 1 | <LOQ | 1.18 ± 0.10 | 78 | 4.62 ± 0.13 | 103 |
| 2 | <LOQ | 1.35 ± 0.03 | 90 | 3.54 ± 0.04 | 79 |
| 3 | <LOQ | 1.55 ± 0.01 | 103 | 4.35 ± 0.09 | 97 |
| 4 | <LOQ | 1.43 ± 0.19 | 95 | 3.97 ± 0.12 | 88 |
| 5 | <LOQ | 1.36 ± 0.01 | 90 | 4.20 ± 0.01 | 93 |
| 6 | <LOQ | 1.32 ± 0.20 | 88 | 3.68 ± 0.23 | 82 |
| 7 | <LOQ | 1.33 ± 0.16 | 89 | 4.14 ± 0.05 | 92 |
| 8 | <LOQ | 1.66 ± 0.09 | 110 | 5.26 ± 0.07 | 117 |
| 9 | <LOQ | 1.57 ± 0.12 | 105 | 4.96 ± 0.13 | 110 |
| 10 | <LOQ | 1.08 ± 0.05 | 72 | 4.02 ± 0.15 | 89 |
| 11 | <LOQ | 1.27 ± 0.10 | 84 | 3.91 ± 0.01 | 87 |
| 12 | <LOQ | 1.77 ± 0.09 | 118 | 4.10 ± 0.10 | 91 |
| 13 | <LOQ | 1.17 ± 0.21 | 78 | 3.96 ± 0.02 | 88 |
| 14 | <LOQ | 1.53 ± 0.14 | 102 | 3.46 ± 0.15 | 77 |
| 15 | <LOQ | 1.29 ± 0.18 | 86 | 4.58 ± 0.10 | 102 |
| 16 | <LOQ | 1.30 ± 0.06 | 87 | 4.20 ± 0.03 | 93 |
| 17 | <LOQ | 1.30 ± 0.30 | 86 | 4.17± 0.21 | 93 |
| 18 | <LOQ | 1.54 ± 0.11 | 102 | 4.77 ± 0.04 | 106 |
| 19 | <LOQ | 1.31 ± 0.02 | 87 | 4.49 ± 0.13 | 100 |
| 20 | <LOQ | 1.82 ± 0.09 | 121 | 4.22 ± 0.20 | 94 |
| 21 | <LOQ | 1.60 ± 0.14 | 107 | 4.19 ± 0.05 | 93 |
| 22 | <LOQ | 1.38 ± 0.25 | 92 | 4.47 ± 0.19 | 99 |
| 23 | <LOQ | 1.48 ± 0.06 | 99 | 4.62 ± 0.01 | 103 |
| 24 | <LOQ | 1.14 ± 0.10 | 76 | 4.19 ± 0.10 | 93 |
| 25 | <LOQ | 1.34 ± 0.19 | 89 | 3.88 ± 0.31 | 86 |
| 26 | <LOQ | 1.30 ± 0.01 | 86 | 3.14 ± 0.04 | 70 |
| 27 | <LOQ | 1.54 ± 0.27 | 103 | 3.89 ± 0.11 | 86 |
| Assay | Unspiked Serum | Spiking 0.80 ng/mL | Spiking 1.60 ng/mL | Spiking 3.10 ng/mL | |||
|---|---|---|---|---|---|---|---|
| Measured (ng/mL) | Recovery % | Measured (ng/mL) | Recovery % | Measured (ng/mL) | Recovery % | ||
| 1 | <LOQ | 0.81 | 102 | 1.51 | 94 | 3.16 | 102 |
| 2 | <LOQ | 0.81 | 102 | 1.52 | 95 | 3.18 | 102 |
| 3 | <LOQ | 0.69 | 86 | 1.47 | 92 | 3.21 | 104 |
| 4 | <LOQ | 0.77 | 96 | 1.54 | 96 | 3.21 | 104 |
| 5 | <LOQ | 0.76 | 95 | 1.51 | 94 | 3.19 | 103 |
| Average | 0.77 | 96 | 1.51 | 94 | 3.2 | 102.8 | |
| CV% | 5.1 | 2.3 | 2.3 | ||||
| Day | Unspiked Serum | Spiking 0.80 ng/mL | Spiking 1.60 ng/mL | Spiking 3.10 ng/mL | |||
|---|---|---|---|---|---|---|---|
| Measured (ng/mL) | Recovery % | Measured (ng/mL) | Recovery % | Measured (ng/mL) | Recovery % | ||
| 1 | <LOQ | 0.77 | 97 | 1.56 | 104 | 3.02 | 97 |
| 2 | <LOQ | 0.67 | 84 | 1.38 | 86 | 2.84 | 92 |
| 3 | <LOQ | 0.65 | 82 | 1.64 | 106 | 3.22 | 104 |
| 4 | <LOQ | 0.58 | 73 | 1.48 | 92 | 2.97 | 96 |
| 5 | <LOQ | 0.77 | 98 | 1.51 | 94 | 3.19 | 103 |
| Average | 0.69 | 86.8 | 1.51 | 96.4 | 3.05 | 98.4 | |
| CV% | 8.1 | 9.8 | 16.0 | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Delfin-Riela, T.; Rossotti, M.; Alvez-Rosado, R.; Leizagoyen, C.; González-Sapienza, G. Highly Sensitive Detection of Zika Virus Nonstructural Protein 1 in Serum Samples by a Two-Site Nanobody ELISA. Biomolecules 2020, 10, 1652. https://doi.org/10.3390/biom10121652
Delfin-Riela T, Rossotti M, Alvez-Rosado R, Leizagoyen C, González-Sapienza G. Highly Sensitive Detection of Zika Virus Nonstructural Protein 1 in Serum Samples by a Two-Site Nanobody ELISA. Biomolecules. 2020; 10(12):1652. https://doi.org/10.3390/biom10121652
Chicago/Turabian StyleDelfin-Riela, Triana, Martín Rossotti, Romina Alvez-Rosado, Carmen Leizagoyen, and Gualberto González-Sapienza. 2020. "Highly Sensitive Detection of Zika Virus Nonstructural Protein 1 in Serum Samples by a Two-Site Nanobody ELISA" Biomolecules 10, no. 12: 1652. https://doi.org/10.3390/biom10121652
APA StyleDelfin-Riela, T., Rossotti, M., Alvez-Rosado, R., Leizagoyen, C., & González-Sapienza, G. (2020). Highly Sensitive Detection of Zika Virus Nonstructural Protein 1 in Serum Samples by a Two-Site Nanobody ELISA. Biomolecules, 10(12), 1652. https://doi.org/10.3390/biom10121652