Systematic Assessment of Protein C-Termini Mutated in Human Disorders
Abstract
:1. Introduction
2. Methods
3. Results
3.1. Rediscoveries of C-Terminal Minimotifs in Disease
3.1.1. CSF1R
3.1.2. FUS
3.1.3. GluN2A
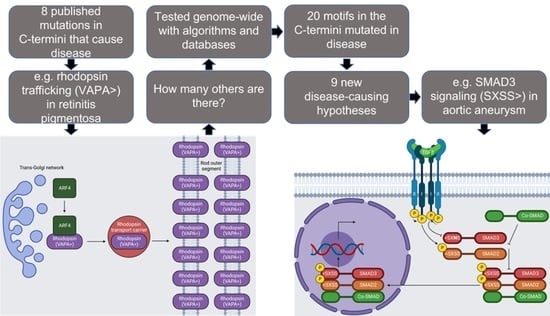
3.1.4. Rhodopsin
3.2. New Hypotheses for C-Terminal Minimotifs in Disease
3.2.1. Androgen Receptor
3.2.2. APOC-III
3.2.3. CRYM
3.2.4. GMPPB
3.2.5. Hemoglobin
3.2.6. SDHA
3.2.7. SMAD3
3.3. Unlikely Hypotheses for C-Terminal Minimotifs in Disease
3.3.1. APOE
3.3.2. INS
4. Discussion
- Twenty missense mutations in PTM sites are reported to cause human disease.
- We developed novel hypotheses for nine mutations in seven genes and PTM sites where disruption of a PTM causes a human disease.
- The analysis of available databases for disease shows that the disruption of C-terminal PTMs, while present, is far less frequent than missense mutations throughout coding regions.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Choi, M.; Scholl, U.I.; Ji, W.; Liu, T.; Tikhonova, I.R.; Zumbo, P.; Nayir, A.; Bakkaloğlu, A.; Özen, S.; Sanjad, S.; et al. Genetic Diagnosis by Whole Exome Capture and Massively Parallel DNA Sequencing. Proc. Natl. Acad. Sci. USA 2009, 106, 19096–19101. [Google Scholar] [CrossRef] [PubMed]
- Barrera, A.; Alastruey-Izquierdo, A.; Martín, M.J.; Cuesta, I.; Vizcaíno, J.A. Analysis of the Protein Domain and Domain Architecture Content in Fungi and Its Application in the Search of New Antifungal Targets. PLoS Comput. Biol. 2014, 10, e1003733. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The Conserved Domain Database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef] [PubMed]
- Sigrist, C.J.A.; de Castro, E.; Cerutti, L.; Cuche, B.A.; Hulo, N.; Bridge, A.; Bougueleret, L.; Xenarios, I. New and Continuing Developments at PROSITE. Nucleic Acids Res. 2012, 41, D344–D347. [Google Scholar] [CrossRef] [PubMed]
- Hulo, N. The PROSITE Database. Nucleic Acids Res. 2006, 34, D227–D230. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, H.; Zhong, H.; Xue, Z. Protein Domain Identification Methods and Online Resources. Comput. Struct. Biotechnol. J. 2021, 19, 1145–1153. [Google Scholar] [CrossRef]
- Lyon, K.F.; Cai, X.; Young, R.J.; Mamun, A.-A.; Rajasekaran, S.; Schiller, M.R. Minimotif Miner 4: A Million Peptide Minimotifs and Counting. Nucleic Acids Res. 2018, 46, D465–D470. [Google Scholar] [CrossRef]
- Balla, S.; Thapar, V.; Verma, S.; Luong, T.; Faghri, T.; Huang, C.-H.; Rajasekaran, S.; del Campo, J.J.; Shinn, J.H.; Mohler, W.A.; et al. Minimotif Miner: A Tool for Investigating Protein Function. Nat Methods 2006, 3, 175–177. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Gouw, M.; Michael, S.; Sámano-Sánchez, H.; Pancsa, R.; Glavina, J.; Diakogianni, A.; Valverde, J.A.; Bukirova, D.; Čalyševa, J.; et al. ELM—The Eukaryotic Linear Motif Resource in 2020. Nucleic Acids Res. 2020, 48, D296–D306. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Toledo, O.; Hedden, M.; Lyon, K.F.; Brooks, S.B.; David, R.P.; Limtong, J.; Newsome, J.M.; Novakovic, N.; Rajasekaran, S.; et al. The Functional Human C-Terminome. PLoS ONE 2016, 11, e0152731. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kadaveru, K. Viral Infection and Human Disease—Insights from Minimotifs. Front. Biosci. 2008, 13, 6455. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Young, R.J.; Chen, J.; Chen, X.; Oh, E.C.; Schiller, M.R. Minimotifs Dysfunction Is Pervasive in Neurodegenerative Disorders. Alzheimer’s Dement. Transl. Res. Clin. Interv. 2018, 4, 414–432. [Google Scholar] [CrossRef] [PubMed]
- Dampier, W.; Evans, P.; Ungar, L.; Tozeren, A. Host Sequence Motifs Shared by HIV Predict Response to Antiretroviral Therapy. BMC Med. Genom. 2009, 2, 47. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Schiller, M.R. The Carboxy-Terminus, a Key Regulator of Protein Function. Crit. Rev. Biochem. Mol. Biol. 2019, 54, 85–102. [Google Scholar] [CrossRef]
- Kupershmidt, S.; Yang, T.; Chanthaphaychith, S.; Wang, Z.; Towbin, J.A.; Roden, D.M. Defective Human Ether-à-Go-Go-Related Gene Trafficking Linked to an Endoplasmic Reticulum Retention Signal in the C Terminus. J. Biol. Chem. 2002, 277, 27442–27448. [Google Scholar] [CrossRef]
- Deretic, D.; Williams, A.H.; Ransom, N.; Morel, V.; Hargrave, P.A.; Arendt, A. Rhodopsin C Terminus, the Site of Mutations Causing Retinal Disease, Regulates Trafficking by Binding to ADP-Ribosylation Factor 4 (ARF4). Proc. Natl. Acad. Sci. USA 2005, 102, 3301–3306. [Google Scholar] [CrossRef]
- Petris, M. A C-Terminal Di-Leucine Is Required for Localization of the Menkes Protein in the Trans-Golgi Network. Hum. Mol. Genet. 1998, 7, 2063–2071. [Google Scholar] [CrossRef]
- Zaarour, N.; Demaretz, S.; Defontaine, N.; Zhu, Y.; Laghmani, K. Multiple Evolutionarily Conserved Di-Leucine Like Motifs in the Carboxyl Terminus Control the Anterograde Trafficking of NKCC2. J. Biol. Chem. 2012, 287, 42642–42653. [Google Scholar] [CrossRef]
- Lam, P.; Xu, S.; Soroka, C.J.; Boyer, J.L. A C-Terminal Tyrosine-Based Motif in the Bile Salt Export Pump Directs Clathrin-Dependent Endocytosis. Hepatology 2012, 55, 1901–1911. [Google Scholar] [CrossRef]
- Reiners, J.; Nagel-Wolfrum, K.; Jürgens, K.; Märker, T.; Wolfrum, U. Molecular Basis of Human Usher Syndrome: Deciphering the Meshes of the Usher Protein Network Provides Insights into the Pathomechanisms of the Usher Disease. Exp. Eye Res. 2006, 83, 97–119. [Google Scholar] [CrossRef]
- Müller, D.; Kausalya, P.J.; Claverie-Martin, F.; Meij, I.C.; Eggert, P.; Garcia-Nieto, V.; Hunziker, W. A Novel Claudin 16 Mutation Associated with Childhood Hypercalciuria Abolishes Binding to ZO-1 and Results in Lysosomal Mistargeting. Am. J. Hum. Genet. 2003, 73, 1293–1301. [Google Scholar] [CrossRef] [PubMed]
- Shy, D.; Gillet, L.; Abriel, H. Cardiac Sodium Channel NaV1.5 Distribution in Myocytes via Interacting Proteins: The Multiple Pool Model. Biochim. Et Biophys. Acta (BBA) Mol. Cell Res. 2013, 1833, 886–894. [Google Scholar] [CrossRef]
- Landrum, M.J.; Lee, J.M.; Benson, M.; Brown, G.; Chao, C.; Chitipiralla, S.; Gu, B.; Hart, J.; Hoffman, D.; Hoover, J.; et al. ClinVar: Public Archive of Interpretations of Clinically Relevant Variants. Nucleic Acids Res. 2016, 44, D862–D868. [Google Scholar] [CrossRef] [PubMed]
- Landrum, M.J.; Chitipiralla, S.; Brown, G.R.; Chen, C.; Gu, B.; Hart, J.; Hoffman, D.; Jang, W.; Kaur, K.; Liu, C.; et al. ClinVar: Improvements to Accessing Data. Nucleic Acids Res. 2020, 48, D835–D844. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Li, C.; Mou, C.; Dong, Y.; Tu, Y. DbNSFP v4: A Comprehensive Database of Transcript-Specific Functional Predictions and Annotations for Human Nonsynonymous and Splice-Site SNVs. Genome Med. 2020, 12, 103. [Google Scholar] [CrossRef]
- Karczewski, K.J.; Francioli, L.C.; Tiao, G.; Cummings, B.B.; Alföldi, J.; Wang, Q.; Collins, R.L.; Laricchia, K.M.; Ganna, A.; Birnbaum, D.P.; et al. The Mutational Constraint Spectrum Quantified from Variation in 141,456 Humans. Nature 2020, 581, 434–443. [Google Scholar] [CrossRef]
- O’Leary, N.A.; Wright, M.W.; Brister, J.R.; Ciufo, S.; Haddad, D.; McVeigh, R.; Rajput, B.; Robbertse, B.; Smith-White, B.; Ako-Adjei, D.; et al. Reference Sequence (RefSeq) Database at NCBI: Current Status, Taxonomic Expansion, and Functional Annotation. Nucleic Acids Res. 2016, 44, D733–D745. [Google Scholar] [CrossRef]
- Bateman, A.; Martin, M.-J.; Orchard, S.; Magrane, M.; Agivetova, R.; Ahmad, S.; Alpi, E.; Bowler-Barnett, E.H.; Britto, R.; The UniProt Consortium; et al. UniProt: The Universal Protein Knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef]
- Database Resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2016, 44, D7–D19. [CrossRef]
- Wajcman, H.; Kister, J.; Marden, M.; Lahary, A.; Monconduit, M.; Galacteros, F. Homoglobin Rouen (α-140 (HC2) Tyr→His): Alteration of the α-Chain C-Terminal Region and Moderate Increase in Oxygen Affinity. Biochim. Et Biophys. Acta (BBA) Mol. Basis Dis. 1992, 1180, 53–57. [Google Scholar] [CrossRef]
- Hornbeck, P.V.; Kornhauser, J.M.; Tkachev, S.; Zhang, B.; Skrzypek, E.; Murray, B.; Latham, V.; Sullivan, M. PhosphoSitePlus: A Comprehensive Resource for Investigating the Structure and Function of Experimentally Determined Post-Translational Modifications in Man and Mouse. Nucleic Acids Res. 2012, 40, D261–D270. [Google Scholar] [CrossRef] [PubMed]
- Zierhut, M.; Haen, S.P.; Moehle, R.; Chan, C.-C. Hematological Neoplasms. In Intraocular Inflammation; Zierhut, M., Pavesio, C., Ohno, S., Orefice, F., Rao, N.A., Eds.; Springer: Berlin/Heidelberg, Germany, 2016; pp. 1493–1510. ISBN 978-3-540-75385-8. [Google Scholar]
- Murga-Zamalloa, C.; Rolland, D.C.M.; Polk, A.; Wolfe, A.; Dewar, H.; Chowdhury, P.; Onder, O.; Dewar, R.; Brown, N.A.; Bailey, N.G.; et al. Colony-Stimulating Factor 1 Receptor (CSF1R) Activates AKT/MTOR Signaling and Promotes T-Cell Lymphoma Viability. Clin. Cancer Res. 2020, 26, 690–703. [Google Scholar] [CrossRef] [PubMed]
- Martín-Moreno, A.M.; Roncador, G.; Maestre, L.; Mata, E.; Jiménez, S.; Martínez-Torrecuadrada, J.L.; Reyes-García, A.I.; Rubio, C.; Tomás, J.F.; Estévez, M.; et al. CSF1R Protein Expression in Reactive Lymphoid Tissues and Lymphoma: Its Relevance in Classical Hodgkin Lymphoma. PLoS ONE 2015, 10, e0125203. [Google Scholar] [CrossRef]
- Ullrich, K.; Wurster, K.D.; Lamprecht, B.; Köchert, K.; Engert, A.; Dörken, B.; Janz, M.; Mathas, S. BAY 43-9006/Sorafenib Blocks CSF1R Activity and Induces Apoptosis in Various Classical Hodgkin Lymphoma Cell Lines. Br. J. Haematol. 2011, 155, 398–402. [Google Scholar] [CrossRef]
- Lamprecht, B.; Walter, K.; Kreher, S.; Kumar, R.; Hummel, M.; Lenze, D.; Köchert, K.; Bouhlel, M.A.; Richter, J.; Soler, E.; et al. Derepression of an Endogenous Long Terminal Repeat Activates the CSF1R Proto-Oncogene in Human Lymphoma. Nat. Med. 2010, 16, 571–579. [Google Scholar] [CrossRef] [PubMed]
- Skinnider, B.F.; Mak, T.W. The Role of Cytokines in Classical Hodgkin Lymphoma. Blood 2002, 99, 4283–4297. [Google Scholar] [CrossRef]
- Komohara, Y.; Noyori, O.; Saito, Y.; Takeya, H.; Baghdadi, M.; Kitagawa, F.; Hama, N.; Ishikawa, K.; Okuno, Y.; Nosaka, K.; et al. Potential Anti-Lymphoma Effect of M-CSFR Inhibitor in Adult T-Cell Leukemia/Lymphoma. JCEH 2018, 58, 152–160. [Google Scholar] [CrossRef]
- Woolford, J.; McAuliffe, A.; Rohrschneider, L.R. Activation of the Feline C-Fms Proto-Oncogene: Multiple Alterations Are Required to Generate a Fully Transformed Phenotype. Cell 1988, 55, 965–977. [Google Scholar] [CrossRef]
- Browning, P.J.; Bunn, H.F.; Cline, A.; Shuman, M.; Nienhuis, A.W. “Replacement” of COOH-Terminal Truncation of v-Fms with c-Fms Sequences Markedly Reduces Transformation Potential. Proc. Natl. Acad. Sci. USA 1986, 83, 7800–7804. [Google Scholar] [CrossRef]
- Ridge, S.A.; Worwood, M.; Oscier, D.; Jacobs, A.; Padua, R.A. FMS Mutations in Myelodysplastic, Leukemic, and Normal Subjects. Proc. Natl. Acad. Sci. USA 1990, 87, 1377–1380. [Google Scholar] [CrossRef] [Green Version]
- Tobal, K.; Pagliuca, A.; Bhatt, B.; Bailey, N.; Layton, D.M.; Mufti, G.J. Mutation of the Human FMS Gene (M-CSF Receptor) in Myelodysplastic Syndromes and Acute Myeloid Leukemia. Leukemia 1990, 4, 486–489. [Google Scholar]
- Yu, W.; Chen, J.; Xiong, Y.; Pixley, F.J.; Dai, X.-M.; Yeung, Y.-G.; Stanley, E.R. CSF-1 Receptor Structure/Function in MacCsf1r-/- Macrophages: Regulation of Proliferation, Differentiation, and Morphology. J. Leukoc. Biol. 2008, 84, 852–863. [Google Scholar] [CrossRef]
- Wilhelmsen, K.; Burkhalter, S.; van der Geer, P. C-Cbl Binds the CSF-1 Receptor at Tyrosine 973, a Novel Phosphorylation Site in the Receptor’s Carboxy-Terminus. Oncogene 2002, 21, 1079–1089. [Google Scholar] [CrossRef] [PubMed]
- Roussel, M.F.; Dull, T.J.; Rettenmier, C.W.; Ralph, P.; Ullrich, A.; Sherr, C.J. Transforming Potential of the C-Fms Proto-Oncogene (CSF-1 Receptor). Nature 1987, 325, 549–552. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Lyashchenko, A.K.; Lu, L.; Nasrabady, S.E.; Elmaleh, M.; Mendelsohn, M.; Nemes, A.; Tapia, J.C.; Mentis, G.Z.; Shneider, N.A. ALS-Associated Mutant FUS Induces Selective Motor Neuron Degeneration through Toxic Gain of Function. Nat. Commun. 2016, 7, 10465. [Google Scholar] [CrossRef]
- Corcia, P.; Danel, V.; Lacour, A.; Beltran, S.; Andres, C.; Couratier, P.; Blasco, H.; Vourc’h, P. A Novel Mutation of the C-Terminal Amino Acid of FUS (Y526C) Strengthens FUS Gene as the Most Frequent Genetic Factor in Aggressive Juvenile ALS. Amyotroph. Lateral Scler. Front. Degener. 2017, 18, 298–301. [Google Scholar] [CrossRef] [PubMed]
- Darovic, S.; Mihevc, S.P.; Župunski, V.; Gunčar, G.; Štalekar, M.; Lee, Y.; Shaw, C.E.; Rogelj, B. Phosphorylation of C-Terminal Tyrosine 526 in FUS Impairs Its Nuclear Import. J. Cell Sci. 2015, 128, 4151–4159. [Google Scholar] [CrossRef]
- Pearl, P.L.; Carrazana, E.J.; Holmes, G.L. The Landau-Kleffner Syndrome. Epilepsy Curr. 2001, 1, 39–45. [Google Scholar] [CrossRef]
- Addis, L.; Virdee, J.K.; Vidler, L.R.; Collier, D.A.; Pal, D.K.; Ursu, D. Epilepsy-Associated GRIN2A Mutations Reduce NMDA Receptor Trafficking and Agonist Potency—Molecular Profiling and Functional Rescue. Sci. Rep. 2017, 7, 66. [Google Scholar] [CrossRef]
- Sapkota, K.; Dore, K.; Tang, K.; Irvine, M.; Fang, G.; Burnell, E.S.; Malinow, R.; Jane, D.E.; Monaghan, D.T. The NMDA Receptor Intracellular C-Terminal Domains Reciprocally Interact with Allosteric Modulators. Biochem. Pharmacol. 2019, 159, 140–153. [Google Scholar] [CrossRef]
- Bowling, K.M.; Thompson, M.L.; Amaral, M.D.; Finnila, C.R.; Hiatt, S.M.; Engel, K.L.; Cochran, J.N.; Brothers, K.B.; East, K.M.; Gray, D.E.; et al. Genomic Diagnosis for Children with Intellectual Disability and/or Developmental Delay. Genome Med. 2017, 9, 43. [Google Scholar] [CrossRef] [Green Version]
- Mota Vieira, M.; Nguyen, T.A.; Wu, K.; Badger, J.D.; Collins, B.M.; Anggono, V.; Lu, W.; Roche, K.W. An Epilepsy-Associated GRIN2A Rare Variant Disrupts CaMKIIα Phosphorylation of GluN2A and NMDA Receptor Trafficking. Cell Rep. 2020, 32, 108104. [Google Scholar] [CrossRef]
- Nash, B.M.; Wright, D.C.; Grigg, J.R.; Bennetts, B.; Jamieson, R.V. Retinal Dystrophies, Genomic Applications in Diagnosis and Prospects for Therapy. Transl. Pediatr. 2015, 4, 139–163. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.E.; Melcher, K.; Xu, H.E. Structure and Activation of Rhodopsin. Acta Pharm. Sin 2012, 33, 291–299. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, M.J.; Lee, K.A.; Niemi, G.A.; Craven, K.B.; Garwin, G.G.; Saari, J.C.; Hurley, J.B. Multiple Phosphorylation of Rhodopsin and the In Vivo Chemistry Underlying Rod Photoreceptor Dark Adaptation. Neuron 2001, 31, 87–101. [Google Scholar] [CrossRef] [PubMed]
- Ohguro, H.; Rudnicka-Nawrot, M.; Buczyko, J.; Zhao, X.; Taylor, J.A.; Walsh, K.A.; Palczewski, K. Structural and Enzymatic Aspects of Rhodopsin Phosphorylation. J. Biol. Chem. 1996, 271, 5215–5224. [Google Scholar] [CrossRef] [PubMed]
- Azevedo, A.W.; Doan, T.; Moaven, H.; Sokal, I.; Baameur, F.; Vishnivetskiy, S.A.; Homan, K.T.; Tesmer, J.J.; Gurevich, V.V.; Chen, J.; et al. C-Terminal Threonines and Serines Play Distinct Roles in the Desensitization of Rhodopsin, a G Protein-Coupled Receptor. eLife 2015, 4, e05981. [Google Scholar] [CrossRef] [PubMed]
- Vishnivetskiy, S.A.; Raman, D.; Wei, J.; Kennedy, M.J.; Hurley, J.B.; Gurevich, V.V. Regulation of Arrestin Binding by Rhodopsin Phosphorylation Level. J. Biol. Chem. 2007, 282, 32075–32083. [Google Scholar] [CrossRef] [PubMed]
- Bownds, D.; Dawes, J.; Miller, J.; Stahlman, M. Phosphorylation of Frog Photoreceptor Membranes Induced by Light. Nat. New Biol. 1972, 237, 125–127. [Google Scholar] [CrossRef] [PubMed]
- Kühn, H.; Dreyer, W.J. Light Dependent Phosphorylation of Rhodopsin by ATP. FEBS Lett. 1972, 20, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Dolph, P.J.; Ranganathan, R.; Colley, N.J.; Hardy, R.W.; Socolich, M.; Zuker, C.S. Arrestin Function in Inactivation of G Protein-Coupled Receptor Rhodopsin in Vivo. Science 1993, 260, 1910–1916. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.S.; Flannery, J.G. Transport of Truncated Rhodopsin and Its Effects on Rod Function and Degeneration. Investig. Ophthalmol. Vis. Sci. 2007, 48, 2868. [Google Scholar] [CrossRef] [PubMed]
- Baylor, D.A.; Burns, M.E. Control of Rhodopsin Activity in Vision. Eye 1998, 12, 521–525. [Google Scholar] [CrossRef] [PubMed]
- Hughes, I.A.; Davies, J.D.; Bunch, T.I.; Pasterski, V.; Mastroyannopoulou, K.; MacDougall, J. Androgen Insensitivity Syndrome. Lancet 2012, 380, 1419–1428. [Google Scholar] [CrossRef]
- Tan, M.E.; Li, J.; Xu, H.E.; Melcher, K.; Yong, E. Androgen Receptor: Structure, Role in Prostate Cancer and Drug Discovery. Acta Pharm. Sin 2015, 36, 3–23. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Dai, B.; Jiang, T.; Xu, K.; Xie, Y.; Kim, O.; Nesheiwat, I.; Kong, X.; Melamed, J.; Handratta, V.D.; et al. Regulation of Androgen Receptor Activity by Tyrosine Phosphorylation. Cancer Cell 2006, 10, 309–319. [Google Scholar] [CrossRef]
- Jenster, G.; van der Korput, H.A.G.M.; van Vroonhoven, C.; van der Kwast, T.H.; Trapman, J.; Brinkmann, A.O. Domains of the Human Androgen Receptor Involved in Steroid Binding, Transcriptional Activation, and Subcellular Localization. Mol. Endocrinol. 1991, 5, 1396–1404. [Google Scholar] [CrossRef]
- Gottlieb, B.; Beitel, L.K.; Nadarajah, A.; Paliouras, M.; Trifiro, M. The Androgen Receptor Gene Mutations Database: 2012 Update. Hum. Mutat. 2012, 33, 887–894. [Google Scholar] [CrossRef]
- Tahiri, B.; Auzou, G.; Nicolas, J.-C.; Sultan, C.; Lupo, B. Participation of Critical Residues from the Extreme C-Terminal End of the Human Androgen Receptor in the Ligand Binding Function. Biochemistry 2001, 40, 8431–8437. [Google Scholar] [CrossRef]
- Fredenrich, A.; Giroux, L.M.; Tremblay, M.; Krimbou, L.; Davignon, J.; Cohn, J.S. Plasma Lipoprotein Distribution of ApoC-III in Normolipidemic and Hypertriglyceridemic Subjects: Comparison of the ApoC-III to ApoE Ratio in Different Lipoprotein Fractions. J. Lipid Res. 1997, 38, 1421–1432. [Google Scholar] [CrossRef]
- Borén, J.; Packard, C.J.; Taskinen, M.-R. The Roles of ApoC-III on the Metabolism of Triglyceride-Rich Lipoproteins in Humans. Front. Endocrinol. 2020, 11, 474. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, M.; Rehues, P.; Iranzo, V.; Mora, J.; Balsells, C.; Guardiola, M.; Ribalta, J. Distribution of Seven ApoC-III Glycoforms in Plasma, VLDL, IDL, LDL and HDL of Healthy Subjects. J. Proteom. 2022, 251, 104398. [Google Scholar] [CrossRef] [PubMed]
- Maeda, H.; Hashimoto, R.K.; Ogura, T.; Hiraga, S.; Uzawa, H. Molecular Cloning of a Human ApoC-III Variant: Thr 74----Ala 74 Mutation Prevents O-Glycosylation. J Lipid Res 1987, 28, 1405–1409. [Google Scholar] [CrossRef] [PubMed]
- Maeda, H.; Uzawa, H.; Kamei, R. Unusual Familial Lipoprotein C-III Associated with Apolipoprotein C-III-0 Preponderance. Biochim. Et Biophys. Acta (BBA) Lipids Lipid Metab. 1981, 665, 578–585. [Google Scholar] [CrossRef]
- Jian, W.; Edom, R.W.; Wang, D.; Weng, N.; Zhang, S. (Weihua) Relative Quantitation of Glycoisoforms of Intact Apolipoprotein C3 in Human Plasma by Liquid Chromatography–High-Resolution Mass Spectrometry. Anal. Chem. 2013, 85, 2867–2874. [Google Scholar] [CrossRef]
- Yassine, H.N.; Trenchevska, O.; Ramrakhiani, A.; Parekh, A.; Koska, J.; Walker, R.W.; Billheimer, D.; Reaven, P.D.; Yen, F.T.; Nelson, R.W.; et al. The Association of Human Apolipoprotein C-III Sialylation Proteoforms with Plasma Triglycerides. PLoS ONE 2015, 10, e0144138. [Google Scholar] [CrossRef]
- Roghani, A.; Zannis, V.I. Mutagenesis of the Glycosylation Site of Human ApoCIII. O-Linked Glycosylation Is Not Required for ApoCIII Secretion and Lipid Binding. J. Biol. Chem. 1988, 263, 17925–17932. [Google Scholar] [CrossRef]
- Savinova, O.V.; Fillaus, K.; Jing, L.; Harris, W.S.; Shearer, G.C. Reduced Apolipoprotein Glycosylation in Patients with the Metabolic Syndrome. PLoS ONE 2014, 9, e104833. [Google Scholar] [CrossRef] [PubMed]
- Tertov, V.V.; Sobenin, I.A.; Tonevitsky, A.G.; Orekhov, A.N.; Smirnov, V.N. Isolation of Atherogenic Modified (Desialylated) Low Density Lipoprotein from Blood of Atherosclerotic Patients: Separation from Native Lipoprotein by Affinity Chromatography. Biochem. Biophys. Res. Commun. 1990, 167, 1122–1127. [Google Scholar] [CrossRef]
- Orekhov, A.N.; Tertov, V.V.; Mukhin, D.N.; Mikhailenko, I.A. Modification of Low Density Lipoprotein by Desialylation Causes Lipid Accumulation in Cultured Cells: Discovery of Desialylated Lipoprotein with Altered Cellular Metabolism in the Blood of Atherosclerotic Patients. Biochem. Biophys. Res. Commun. 1989, 162, 206–211. [Google Scholar] [CrossRef]
- Tertov, V.V.; Orekhov, A.N.; Sobenin, I.A.; Morrisett, J.D.; Gotto, A.M.; Guevara, J.G. Carbohydrate Composition of Protein and Lipid Components in Sialic Acid-Rich and -Poor Low Density Lipoproteins from Subjects with and without Coronary Artery Disease. J. Lipid Res. 1993, 34, 365–375. [Google Scholar] [CrossRef]
- Millar, J.S.; Anber, V.; Shepherd, J.; Packard, C.J. Sialic Acid-Containing Components of Lipoproteins Influence Lipoprotein-Proteoglycan Interactions. Atherosclerosis 1999, 145, 253–260. [Google Scholar] [CrossRef]
- Mendoza, S.; Trenchevska, O.; King, S.M.; Nelson, R.W.; Nedelkov, D.; Krauss, R.M.; Yassine, H.N. Changes in Low-Density Lipoprotein Size Phenotypes Associate with Changes in Apolipoprotein C-III Glycoforms after Dietary Interventions. J. Clin. Lipidol. 2017, 11, 224–233.e2. [Google Scholar] [CrossRef]
- Koska, J.; Yassine, H.; Trenchevska, O.; Sinari, S.; Schwenke, D.C.; Yen, F.T.; Billheimer, D.; Nelson, R.W.; Nedelkov, D.; Reaven, P.D. Disialylated Apolipoprotein C-III Proteoform Is Associated with Improved Lipids in Prediabetes and Type 2 Diabetes. J. Lipid Res. 2016, 57, 894–905. [Google Scholar] [CrossRef]
- Krishnan, S.; Shimoda, M.; Sacchi, R.; Kailemia, M.J.; Luxardi, G.; Kaysen, G.A.; Parikh, A.N.; Ngassam, V.N.; Johansen, K.; Chertow, G.M.; et al. HDL Glycoprotein Composition and Site-Specific Glycosylation Differentiates Between Clinical Groups and Affects IL-6 Secretion in Lipopolysaccharide-Stimulated Monocytes. Sci. Rep. 2017, 7, 43728. [Google Scholar] [CrossRef]
- Kashyap, M.L.; Srivastava, L.S.; Hynd, B.A.; Gartside, P.S.; Perisutti, G. Quantitation of Human Apolipoprotein C-III and Its Subspecie by Radioimmunoassay and Analytical Isoelectric Focusing: Abnormal Plasma Triglyceride-Rich Lipoprotein Apolipoprotein C-III Subspecie Concentrations in Hypertriglyceridemia. J. Lipid Res. 1981, 22, 800–810. [Google Scholar] [CrossRef] [PubMed]
- Holdsworth, G.; Stocks, J.; Dodson, P.; Galton, D.J. An Abnormal Triglyceride-Rich Lipoprotein Containing Excess Sialylated Apolipoprotein C-III. J. Clin. Investig. 1982, 69, 932–939. [Google Scholar] [CrossRef]
- Holleboom, A.G.; Karlsson, H.; Lin, R.-S.; Beres, T.M.; Sierts, J.A.; Herman, D.S.; Stroes, E.S.G.; Aerts, J.M.; Kastelein, J.J.P.; Motazacker, M.M.; et al. Heterozygosity for a Loss-of-Function Mutation in GALNT2 Improves Plasma Triglyceride Clearance in Man. Cell Metab. 2011, 14, 811–818. [Google Scholar] [CrossRef] [PubMed]
- Abe, S.; Katagiri, T.; Saito-Hisaminato, A.; Usami, S.; Inoue, Y.; Tsunoda, T.; Nakamura, Y. Identification of CRYM as a Candidate Responsible for Nonsyndromic Deafness, through CDNA Microarray Analysis of Human Cochlear and Vestibular Tissues**Nucleotide Sequence Data Reported Herein Are Available in the DDBJ/EMBL/GenBank Databases; for Details, See the Electronic-Database Information Section of This Article. Am. J. Hum. Genet. 2003, 72, 73–82. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Li, Q.; Deng, A.; Zhu, X.; Yang, J. Identification of a Novel Mutation in CRYM in a Chinese Family with Hearing Loss Using Whole-exome Sequencing. Exp. Ther. Med. 2020, 20, 1447–1454. [Google Scholar] [CrossRef]
- Kinney, C.J.; Bloch, R.J. Μ-Crystallin: A Thyroid Hormone Binding Protein. Endocr. Regul. 2021, 55, 89–102. [Google Scholar] [CrossRef]
- Oshima, A. CRYM Mutations Cause Deafness through Thyroid Hormone Binding Properties in the Fibrocytes of the Cochlea. J. Med. Genet. 2006, 43, e25. [Google Scholar] [CrossRef]
- Carss, K.J.; Stevens, E.; Foley, A.R.; Cirak, S.; Riemersma, M.; Torelli, S.; Hoischen, A.; Willer, T.; van Scherpenzeel, M.; Moore, S.A.; et al. Mutations in GDP-Mannose Pyrophosphorylase B Cause Congenital and Limb-Girdle Muscular Dystrophies Associated with Hypoglycosylation of α-Dystroglycan. Am. J. Hum. Genet. 2013, 93, 29–41. [Google Scholar] [CrossRef] [PubMed]
- Luo, S.; Cai, S.; Maxwell, S.; Yue, D.; Zhu, W.; Qiao, K.; Zhu, Z.; Zhou, L.; Xi, J.; Lu, J.; et al. Novel Mutations in the C-Terminal Region of GMPPB Causing Limb-Girdle Muscular Dystrophy Overlapping with Congenital Myasthenic Syndrome. Neuromuscul. Disord. 2017, 27, 557–564. [Google Scholar] [CrossRef]
- Sun, L.; Shen, D.; Xiong, T.; Zhou, Z.; Lu, X.; Cui, F. Limb-Girdle Muscular Dystrophy Due to GMPPB Mutations: A Case Report and Comprehensive Literature Review. Bosn J. Basic. Med. Sci. 2020, 2, 275. [Google Scholar] [CrossRef]
- Liu, Z.; Wang, Y.; Yang, F.; Yang, Q.; Mo, X.; Burstein, E.; Jia, D.; Cai, X.; Tu, Y. GMPPB-Congenital Disorders of Glycosylation Associate with Decreased Enzymatic Activity of GMPPB. Mol. Biomed. 2021, 2, 13. [Google Scholar] [CrossRef]
- Zheng, L.; Liu, Z.; Wang, Y.; Yang, F.; Wang, J.; Huang, W.; Qin, J.; Tian, M.; Cai, X.; Liu, X.; et al. Cryo-EM Structures of Human GMPPA–GMPPB Complex Reveal How Cells Maintain GDP-Mannose Homeostasis. Nat. Struct. Mol. Biol. 2021, 28, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Yoshida-Moriguchi, T.; Yu, L.; Stalnaker, S.H.; Davis, S.; Kunz, S.; Madson, M.; Oldstone, M.B.A.; Schachter, H.; Wells, L.; Campbell, K.P. O-Mannosyl Phosphorylation of Alpha-Dystroglycan Is Required for Laminin Binding. Science 2010, 327, 88–92. [Google Scholar] [CrossRef]
- Endo, T.; Toda, T. Glycosylation in Congenital Muscular Dystrophies. Biol. Pharm. Bull. 2003, 26, 1641–1647. [Google Scholar] [CrossRef]
- Belaya, K.; Rodríguez Cruz, P.M.; Liu, W.W.; Maxwell, S.; McGowan, S.; Farrugia, M.E.; Petty, R.; Walls, T.J.; Sedghi, M.; Basiri, K.; et al. Mutations in GMPPB Cause Congenital Myasthenic Syndrome and Bridge Myasthenic Disorders with Dystroglycanopathies. Brain 2015, 138, 2493–2504. [Google Scholar] [CrossRef]
- Bento, C. Genetic Basis of Congenital Erythrocytosis. Int. J. Lab. Hem. 2018, 40, 62–67. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, M.H.; Ghatge, M.S.; Safo, M.K. Hemoglobin: Structure, Function and Allostery. In Vertebrate and Invertebrate Respiratory Proteins, Lipoproteins and other Body Fluid Proteins; Hoeger, U., Harris, J.R., Eds.; Subcellular Biochemistry; Springer International Publishing: Cham, Switzerland, 2020; Volume 94, pp. 345–382. ISBN 978-3-030-41768-0. [Google Scholar]
- Wajcman, H.; Galactéros, F. Hemoglobins with High Oxygen Affinity Leading to Erythrocytosis. New Variants and New Concepts. LHEM 2005, 29, 91–106. [Google Scholar] [CrossRef]
- Hardison, R.C.; Chui, D.H.K.; Giardine, B.; Riemer, C.; Patrinos, G.P.; Anagnou, N.; Miller, W.; Wajcman, H. HbVar: A Relational Database of Human Hemoglobin Variants and Thalassemia Mutations at the Globin Gene Server. Hum. Mutat. 2002, 19, 225–233. [Google Scholar] [CrossRef] [PubMed]
- Giardine, B.; van Baal, S.; Kaimakis, P.; Riemer, C.; Miller, W.; Samara, M.; Kollia, P.; Anagnou, N.P.; Chui, D.H.K.; Wajcman, H.; et al. HbVar Database of Human Hemoglobin Variants and Thalassemia Mutations: 2007 Update. Hum. Mutat. 2007, 28, 206. [Google Scholar] [CrossRef] [PubMed]
- Perutz, M.F.; Rossmann, M.G.; Cullis, A.F.; Muirhead, H.; Will, G.; North, A.C.T. Structure of Hæmoglobin: A Three-Dimensional Fourier Synthesis at 5.5-Å. Resolution, Obtained by X-Ray Analysis. Nature 1960, 185, 416–422. [Google Scholar] [CrossRef]
- Perutz, M.F.; Muirhead, H.; Cox, J.M.; Goaman, L.C.G. Three-Dimensional Fourier Synthesis of Horse Oxyhaemoglobin at 2.8 Å Resolution: The Atomic Model. Nature 1968, 219, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Perutz, M.F. Stereochemistry of Cooperative Effects in Haemoglobin: Haem–Haem Interaction and the Problem of Allostery. Nature 1970, 228, 726–734. [Google Scholar] [CrossRef]
- Kneipp, J.; Balakrishnan, G.; Chen, R.; Shen, T.-J.; Sahu, S.C.; Ho, N.T.; Giovannelli, J.L.; Simplaceanu, V.; Ho, C.; Spiro, T.G. Dynamics of Allostery in Hemoglobin: Roles of the Penultimate Tyrosine H Bonds. J. Mol. Biol. 2006, 356, 335–353. [Google Scholar] [CrossRef] [PubMed]
- Rajko Igić, B.M.; Leray, V.; Deddish, P.A.; Erdös, E.G. Removal of Arg 141 From the α Chain of Human Hemoglobin by Carboxypeptidases N and M. Circ. Res. 1996, 78, 635–642. [Google Scholar] [CrossRef]
- Tosqui, P.; Bonini-Domingos, C.R.; Colombo, M.F. Characterization and Oxygen Binding Properties of Des-Arg Human Hemoglobin. Braz J. Med. Biol. Res. 2009, 42, 494–500. [Google Scholar] [CrossRef]
- Bare, G.H.; Bromberg, P.A.; Alben, J.O.; Brimhall, B.; Jones, R.T.; Mintz, S.; Rother, I. Altered C-Terminal Salt Bridges in Haemoglobin York Cause High Oxygen Affinity. Nature 1976, 259, 155–156. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Spiro, T.G. Structure Changes in Hemoglobin upon Deletion of C-Terminal Residues, Monitored by Resonance Raman Spectroscopy. Biochemistry 1998, 37, 9940–9951. [Google Scholar] [CrossRef]
- Antonini, E.; Wyman, J.; Zito, R.; Rossi-Fanelli, A.; Caputo, A. Studies on Carboxypeptidase Digests of Human Hemoglobin. J. Biol. Chem. 1961, 236, PC60–PC63. [Google Scholar] [CrossRef] [PubMed]
- Bettati, S.; Mozzarelli, A.; Perutz, M.F. Allosteric Mechanism of Haemoglobin: Rupture of Salt-Bridges Raises the Oxygen Affinity of the T-Structure 1 1Edited by D. Rees. J. Mol. Biol. 1998, 281, 581–585. [Google Scholar] [CrossRef] [PubMed]
- Jones, R.T.; Shih, T.-B. Hemoglobin Variants with Altered Oxygen Affinity. Hemoglobin 1980, 4, 243–261. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, R.M.; Jue, D.L.; Ali, M.A.M.; Lyonnais, J.; Moo-Penn, W.F. Hemoglobiri Bethesda, Β145 (HC2) Tyr→His, in a Canadian Family. Am J Clin Pathol 1976, 66, 449–452. [Google Scholar] [CrossRef]
- Adamson, J.W.; Hayashi, A.; Stamatoyannopoulos, G.; Burger, W.F. Erythrocyte Function and Marrow Regulation in Hemoglobin Bethesda (Β145 Histidine). J. Clin. Investig. 1972, 51, 2883–2888. [Google Scholar] [CrossRef]
- Bunn, H.F.; Bradley, T.B.; Davis, W.E.; Drysdale, J.W.; Burke, J.F.; Beck, W.S.; Laver, M.B. Structural and Functional Studies on Hemoglobin Bethesda (A2β2145 His), a Variant Associated with Compensatory Erythrocytosis. J. Clin. Investig. 1972, 51, 2299–2309. [Google Scholar] [CrossRef]
- Hayashi, A.; Stamatoyannopoulos, G.; Yoshida, A.; Adamson, J. Haemoglobin Rainier: Β145 (HC2) Tyrosine→Cysteine and Haemoglobin Bethesda: Β145 (HC2) Tyrosine → Histidine. Nat. New Biol. 1971, 230, 264–267. [Google Scholar] [CrossRef]
- Gacon, G.; Wajcman, H.; Labie, D.; Vigneron, C. Structural and Functional Study of Hb Nancy β 145 (HC 2) Tyr → Asp a High Oxygen Affinity Hemoglobin. FEBS Lett. 1975, 56, 39–42. [Google Scholar] [CrossRef]
- Charache, S.; Brimhall, B.; Jones, R.T. Polycythemia Produced by Hemoglobin Osler (Beta-145 (HC2) Tyr Yields Asp). Johns Hopkins Med. J. 1975, 136, 132–136. [Google Scholar] [PubMed]
- Hutt, P.J.; Donaldson, M.H.; Khatri, J.; Fairbanks, V.F.; Hoyer, J.D.; Thibodeau, S.N.; Moxness, M.S.; McMorrow, L.E.; Green, M.M.; Jones, R.T. Hemoglobin S/Hemoglobin Osler: A Case with 3 β Globin Chains. DNA Sequence (AAT) Proves That Hb Osler Is β 145 Tyr→Asn. Am. J. Hematol. 1996, 52, 305–309. [Google Scholar] [CrossRef]
- Hebbel, R.P.; Eaton, J.W.; Kronenberg, R.S.; Zanjani, E.D.; Moore, L.G.; Berger, E.M. Human Llamas: Adaptation to Altitude in Subjects with High Hemoglobin Oxygen Affinity. J. Clin. Investig. 1978, 62, 593–600. [Google Scholar] [CrossRef]
- Zak, S.J.; Brimhall, B.; Jones, R.T.; Kaplan, M.E. Hemoglobin Andrew-Minneapolis Alpha 2 A Beta 2 144 Lys Leads to Asn: A New High-Oxygen-Affinity Mutant Human Hemoglobin. Blood 1974, 44, 543–549. [Google Scholar] [CrossRef]
- Rahbar, S.; Lee, T.D.; Davis, M.; Novotny, W.F.; Ranney, H.M. A Second Case of Hb Hanamaki [α 2 139 (HC1) LYS→GLU β 2 ] in an American Family with Erythrocytosis. Hemoglobin 1994, 18, 221–226. [Google Scholar] [CrossRef] [PubMed]
- Orisaka, M.; Tajima, T.; Harano, T.; Harano, K.; Kushida, Y.; Imai, K. A New α Chain Variant, HB Hanamaki or α 2 139(HC1)LYS→GLUβ 2, Found in a Japanese Family. Hemoglobin 1992, 16, 67–71. [Google Scholar] [CrossRef]
- Klotz, I.M.; Tam, J.W.O. Acetylation of Sickle Cell Hemoglobin by Aspirin. Proc. Natl. Acad. Sci. USA 1973, 70, 1313–1315. [Google Scholar] [CrossRef] [PubMed]
- Shamsuddin, M.; Mason, R.G.; Ritchey, J.M.; Honig, G.R.; Klotz, I.M. Sites of Acetylation of Sickle Cell Hemoglobin by Aspirin. Proc. Natl. Acad. Sci. USA 1974, 71, 4693–4697. [Google Scholar] [CrossRef]
- de Furia, F.G.; Cerami, A.; Bunn, H.F.; Lu, Y.S.; Peterson, C.M. The Effect of Aspirin on Sickling and Oxygen Affinity of Erythrocytes. Proc. Natl. Acad. Sci. USA 1973, 70, 3707–3710. [Google Scholar] [CrossRef]
- Webber, B.B.; Wilson, J.B.; Gu, L.-H.; Huisman, T.H.J. HB Ethiopia or α 2 140(HC2)TYR→HISβ 2. Hemoglobin 1992, 16, 441–443. [Google Scholar] [CrossRef]
- Fullerton, M.; McFarland, R.; Taylor, R.W.; Alston, C.L. The Genetic Basis of Isolated Mitochondrial Complex II Deficiency. Mol. Genet. Metab. 2020, 131, 53–65. [Google Scholar] [CrossRef]
- Renkema, G.H.; Wortmann, S.B.; Smeets, R.J.; Venselaar, H.; Antoine, M.; Visser, G.; Ben-Omran, T.; van den Heuvel, L.P.; Timmers, H.J.L.M.; Smeitink, J.A.; et al. SDHA Mutations Causing a Multisystem Mitochondrial Disease: Novel Mutations and Genetic Overlap with Hereditary Tumors. Eur. J. Hum. Genet 2015, 23, 202–209. [Google Scholar] [CrossRef]
- Burnichon, N.; Brière, J.-J.; Libé, R.; Vescovo, L.; Rivière, J.; Tissier, F.; Jouanno, E.; Jeunemaitre, X.; Bénit, P.; Tzagoloff, A.; et al. SDHA Is a Tumor Suppressor Gene Causing Paraganglioma. Hum. Mol. Genet. 2010, 19, 3011–3020. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Maklashina, E.; Cecchini, G.; Iverson, T.M. The Roles of SDHAF2 and Dicarboxylate in Covalent Flavinylation of SDHA, the Human Complex II Flavoprotein. Proc. Natl. Acad. Sci. USA 2020, 117, 23548–23556. [Google Scholar] [CrossRef]
- Hao, H.-X.; Khalimonchuk, O.; Schraders, M.; Dephoure, N.; Bayley, J.-P.; Kunst, H.; Devilee, P.; Cremers, C.W.R.J.; Schiffman, J.D.; Bentz, B.G.; et al. SDH5, a Gene Required for Flavination of Succinate Dehydrogenase, Is Mutated in Paraganglioma. Science 2009, 325, 1139–1142. [Google Scholar] [CrossRef]
- Rutter, J.; Winge, D.R.; Schiffman, J.D. Succinate Dehydrogenase—Assembly, Regulation and Role in Human Disease. Mitochondrion 2010, 10, 393–401. [Google Scholar] [CrossRef] [PubMed]
- Zehavi, Y.; Saada, A.; Jabaly-Habib, H.; Dessau, M.; Shaag, A.; Elpeleg, O.; Spiegel, R. A Novel de Novo Heterozygous Pathogenic Variant in the SDHA Gene Results in Childhood Onset Bilateral Optic Atrophy and Cognitive Impairment. Metab. Brain Dis. 2021, 36, 581–588. [Google Scholar] [CrossRef]
- Kim, H.J.; Jeong, M.-Y.; Na, U.; Winge, D.R. Flavinylation and Assembly of Succinate Dehydrogenase Are Dependent on the C-Terminal Tail of the Flavoprotein Subunit. J. Biol. Chem. 2012, 287, 40670–40679. [Google Scholar] [CrossRef]
- Jones, J.A.; Spinale, F.G.; Ikonomidis, J.S. Transforming Growth Factor-β Signaling in Thoracic Aortic Aneurysm Development: A Paradox in Pathogenesis. J. Vasc. Res. 2009, 46, 119–137. [Google Scholar] [CrossRef]
- Goldfinger, J.Z.; Halperin, J.L.; Marin, M.L.; Stewart, A.S.; Eagle, K.A.; Fuster, V. Thoracic Aortic Aneurysm and Dissection. J. Am. Coll. Cardiol. 2014, 64, 1725–1739. [Google Scholar] [CrossRef]
- Regalado, E.S.; Guo, D.; Villamizar, C.; Avidan, N.; Gilchrist, D.; McGillivray, B.; Clarke, L.; Bernier, F.; Santos-Cortez, R.L.; Leal, S.M.; et al. Exome Sequencing Identifies SMAD3 Mutations as a Cause of Familial Thoracic Aortic Aneurysm and Dissection With Intracranial and Other Arterial Aneurysms. Circ. Res. 2011, 109, 680–686. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van der Pluijm, I.; van Vliet, N.; von der Thusen, J.H.; Robertus, J.L.; Ridwan, Y.; van Heijningen, P.M.; van Thiel, B.S.; Vermeij, M.; Hoeks, S.E.; Buijs-Offerman, R.M.G.B.; et al. Defective Connective Tissue Remodeling in Smad3 Mice Leads to Accelerated Aneurysmal Growth Through Disturbed Downstream TGF-β Signaling. EBioMedicine 2016, 12, 280–294. [Google Scholar] [CrossRef] [PubMed]
- Schepers, D.; Tortora, G.; Morisaki, H.; MacCarrick, G.; Lindsay, M.; Liang, D.; Mehta, S.G.; Hague, J.; Verhagen, J.; van de Laar, I.; et al. A Mutation Update on the LDS-Associated Genes TGFB2/3 and SMAD2/3. Hum. Mutat. 2018, 39, 621–634. [Google Scholar] [CrossRef] [PubMed]
- Hostetler, E.M.; Regalado, E.S.; Guo, D.-C.; Hanna, N.; Arnaud, P.; Muiño-Mosquera, L.; Callewaert, B.L.; Lee, K.; Leal, S.M.; Wallace, S.E.; et al. SMAD3 Pathogenic Variants: Risk for Thoracic Aortic Disease and Associated Complications from the Montalcino Aortic Consortium. J. Med. Genet 2019, 56, 252–260. [Google Scholar] [CrossRef]
- Brown, K.A.; Pietenpol, J.A.; Moses, H.L. A Tale of Two Proteins: Differential Roles and Regulation of Smad2 and Smad3 in TGF-β Signaling. J. Cell. Biochem. 2007, 101, 9–33. [Google Scholar] [CrossRef]
- Flowers, S.A.; Rebeck, G.W. APOE in the Normal Brain. Neurobiol. Dis. 2020, 136, 104724. [Google Scholar] [CrossRef]
- Kim, J.; Basak, J.M.; Holtzman, D.M. The Role of Apolipoprotein E in Alzheimer’s Disease. Neuron 2009, 63, 287–303. [Google Scholar] [CrossRef] [PubMed]
- van den Maagdenberg, A.M.; Weng, W.; de Bruijn, I.H.; de Knijff, P.; Funke, H.; Smelt, A.H.; Gevers Leuven, J.A.; van’t Hooft, F.M.; Assmann, G.; Hofker, M.H. Characterization of Five New Mutants in the Carboxyl-Terminal Domain of Human Apolipoprotein E: No Cosegregation with Severe Hyperlipidemia. Am. J. Hum. Genet 1993, 52, 937–946. [Google Scholar]
- Halim, A.; Rüetschi, U.; Larson, G.; Nilsson, J. LC–MS/MS Characterization of O-Glycosylation Sites and Glycan Structures of Human Cerebrospinal Fluid Glycoproteins. J. Proteome Res. 2013, 12, 573–584. [Google Scholar] [CrossRef]
- Raftery, M.; Campbell, R.; Glaros, E.N.; Rye, K.-A.; Halliday, G.M.; Jessup, W.; Garner, B. Phosphorylation of Apolipoprotein-E at an Atypical Protein Kinase CK2 PSD/E Site in Vitro. Biochemistry 2005, 44, 7346–7353. [Google Scholar] [CrossRef]
- Hu, Y.; Meuret, C.; Go, S.; Yassine, H.N.; Nedelkov, D. Simple and Fast Assay for Apolipoprotein E Phenotyping and Glycotyping: Discovering Isoform-Specific Glycosylation in Plasma and Cerebrospinal Fluid. JAD 2020, 76, 883–893. [Google Scholar] [CrossRef] [PubMed]
- Chua, C.-C.; Lim, M.-L.; Wong, B.-S. Altered Apolipoprotein E Glycosylation Is Associated with Aβ(42) Accumulation in an Animal Model of Niemann-Pick Type C Disease. J. Neurochem. 2010, 112, 1619–1626. [Google Scholar] [CrossRef] [PubMed]
- Sugano, M.; Yamauchi, K.; Kawasaki, K.; Tozuka, M.; Fujita, K.; Okumura, N.; Ota, H. Sialic Acid Moiety of Apolipoprotein E3 at Thr194 Affects Its Interaction with β-Amyloid1–42 Peptides. Clin. Chim. Acta 2008, 388, 123–129. [Google Scholar] [CrossRef] [PubMed]
- Ke, L.; Chan, H.; Chen, C.; Chang, C.; Lu, P.; Chu, C.; Lai, W.; Shin, S.; Liu, F.; Chen, C. Increased APOE Glycosylation Plays a Key Role in the Atherogenicity of L5 Low-density Lipoprotein. FASEB J. 2020, 34, 9802–9813. [Google Scholar] [CrossRef]
- Wernette-Hammond, M.E.; Lauer, S.J.; Corsini, A.; Walker, D.; Taylor, J.M.; Rall, S.C. Glycosylation of Human Apolipoprotein E. The Carbohydrate Attachment Site Is Threonine 194. J. Biol. Chem. 1989, 264, 9094–9101. [Google Scholar] [CrossRef]
- Zanni, E.E.; Kouvatsi, A.; Hadzopoulou-Cladaras, M.; Krieger, M.; Zannis, V.I. Expression of ApoE Gene in Chinese Hamster Cells with a Reversible Defect in O-Glycosylation. Glycosylation Is Not Required for ApoE Secretion. J. Biol. Chem. 1989, 264, 9137–9140. [Google Scholar] [CrossRef]
- Edghill, E.L.; Flanagan, S.E.; Patch, A.-M.; Boustred, C.; Parrish, A.; Shields, B.; Shepherd, M.H.; Hussain, K.; Kapoor, R.R.; Malecki, M.; et al. Insulin Mutation Screening in 1044 Patients With Diabetes. Diabetes 2008, 57, 1034–1042. [Google Scholar] [CrossRef]
- Adhikari, S.; Nice, E.C.; Deutsch, E.W.; Lane, L.; Omenn, G.S.; Pennington, S.R.; Paik, Y.-K.; Overall, C.M.; Corrales, F.J.; Cristea, I.M.; et al. A High-Stringency Blueprint of the Human Proteome. Nat. Commun. 2020, 11, 5301. [Google Scholar] [CrossRef]
- Morgan, A.A.; Rubenstein, E. Proline: The Distribution, Frequency, Positioning, and Common Functional Roles of Proline and Polyproline Sequences in the Human Proteome. PLoS ONE 2013, 8, e53785. [Google Scholar] [CrossRef]
- Davydov, E.V.; Goode, D.L.; Sirota, M.; Cooper, G.M.; Sidow, A.; Batzoglou, S. Identifying a High Fraction of the Human Genome to Be under Selective Constraint Using GERP++. PLoS Comput. Biol. 2010, 6, e1001025. [Google Scholar] [CrossRef]
- Ng, P.C. SIFT: Predicting Amino Acid Changes That Affect Protein Function. Nucleic Acids Res. 2003, 31, 3812–3814. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Henikoff, S.; Ng, P.C. Predicting the Effects of Coding Non-Synonymous Variants on Protein Function Using the SIFT Algorithm. Nat Protoc 2009, 4, 1073–1081. [Google Scholar] [CrossRef] [PubMed]
- Vaser, R.; Adusumalli, S.; Leng, S.N.; Sikic, M.; Ng, P.C. SIFT Missense Predictions for Genomes. Nat. Protoc. 2016, 11, 1–9. [Google Scholar] [CrossRef]
- Adzhubei, I.; Jordan, D.M.; Sunyaev, S.R. Predicting Functional Effect of Human Missense Mutations Using PolyPhen-2. CP Hum. Genet. 2013, 76, 7–20. [Google Scholar] [CrossRef]
- Samocha, K.E.; Kosmicki, J.A.; Karczewski, K.J.; O’Donnell-Luria, A.H.; Pierce-Hoffman, E.; MacArthur, D.G.; Neale, B.M.; Daly, M.J. Regional Missense Constraint Improves Variant Deleteriousness Prediction. BioRxiv 2017, 148353. [Google Scholar]
- Rentzsch, P.; Witten, D.; Cooper, G.M.; Shendure, J.; Kircher, M. CADD: Predicting the Deleteriousness of Variants throughout the Human Genome. Nucleic Acids Res. 2019, 47, D886–D894. [Google Scholar] [CrossRef]
- Kircher, M.; Witten, D.M.; Jain, P.; O’Roak, B.J.; Cooper, G.M.; Shendure, J. A General Framework for Estimating the Relative Pathogenicity of Human Genetic Variants. Nat. Genet. 2014, 46, 310–315. [Google Scholar] [CrossRef] [Green Version]









| Protein | Minimotif | General Function | Disease | Reference |
|---|---|---|---|---|
| Kv11.1 | RGRX152> 1 | Trafficking | Long QT syndrome type 2 | Kupershmidt et al. (2002), [15] |
| Rhodopsin | VAPA> | Trafficking | Vision loss | Deretic et al. (2005), [16] |
| MNK | LLX12> 1 | Trafficking | Menkes disease | Petris et al. (1998), [17] |
| NKCC2 | LLX13> 1 | Trafficking | Bartter syndrome | Zaarour et al. (2012), [18] |
| BSEP | YYKLVX7> 1 | Trafficking | Primary familial intrahepatic cholestasis type 2 | Lam et al. (2012), [19] |
| SANS | TEL> | Binding | Usher syndrome type 1 | Reiners et al. (2006), [20] |
| Claudin-16 | TRV> | Binding | Nephrocalcinosis | Müller et al. (2003), [21] |
| NaV1.5 | SIV> | Binding | Cardiac disease | Shy et al. (2013), [22] |
| Category | Count |
|---|---|
| Total ClinVar variants | 1,260,173 |
| ClinVar nonsense and C-terminal missense variants | 37,046 |
| ClinVar C-terminal missense variants | 5550 |
| Pathogenic variants with PTM disrupted | 20 |
| Uncertain variants with PTM disrupted | 119 |
| Total with PTM disrupted | 213 |
| Pathogenic with motif but not PTM disrupted | 166 |
| Uncertain with motif but not PTM disrupted | 782 |
| Total with motif but not PTM disrupted | 1457 |
| Total C-terminal motifs | 9165 |
| Motifs mutated by a C-terminal variant | 1258 |
| Protein | Disease | Pathogenicity | Mutation | Motif | PTM | Frequency | GERP Score | Type |
|---|---|---|---|---|---|---|---|---|
| AR | Androgen resistance syndrome | Likely pathogenic | Y915S | KVKPIYFHTQ> | Phosphotyrosine | <7.95 × 106 | 5.18 | New hypothesis |
| APOC-III | NULL 1 | NULL 1 | T74A | PEVRPTSAVAA> | O-glycosylation | 2.78 × 105 | −0.501 | New hypothesis |
| APOE | NULL 2 | NULL 2 | S296R | TSAAPVPSDNH> | O-glycosylation; phosphoserine | 9.48 × 105 | −0.0552 | Unlikely |
| CRYM | Congenital sensorineural deafness | Pathogenic | K314T | YDSWSSGK> | Acetylation | <7.95 × 106 | 5.45 | New hypothesis |
| CSF1R | Hematologic neoplasm | Likely pathogenic | Y969F | LQPNNYQFC> | Phosphotyrosine | <7.95 × 106 | 5.25 | Rediscovery |
| CSF1R | Hematologic neoplasm | Likely pathogenic | Y969H | LQPNNYQFC> | Phosphotyrosine | <7.95 × 106 | 5.25 | Rediscovery |
| CSF1R | Hematologic neoplasm | Likely pathogenic | Y969C | LQPNNYQFC> | Phosphotyrosine | <7.95 × 106 | 5.25 | Rediscovery |
| FUS | Juvenile amyotrophic lateral sclerosis | Pathogenic | Y526C | QDRRERPY> | Phosphotyrosine | 3.98 × 106 | 0.158 | Rediscovery |
| GluN2A | Landau-Kleffner syndrome | Likely pathogenic | S1459G | RVYKKMPSIESDV> | Phosphoserine | <7.95 × 106 | 5.79 | Rediscovery |
| GMPPB | Muscular dystrophy-dystroglycanopathy | Likely pathogenic | R357H | GESVPEPRIIM> | Methylation | 3.72 × 105 | 5.24 | New hypothesis |
| HBA | Familial erythrocytosis | Pathogenic | K139E | VSTVLTSKYR> | Acetylation | <7.95 × 106 | 4.43 | Unlikely |
| HBA | NULL 3 | NULL 3 | Y140H | VSTVLTSKYR> | Phosphotyrosine | <7.95 × 106 | 2.8 | Unlikely |
| HBB | Familial erythrocytosis | Pathogenic | K144N | VANALAHKYH> | Acetylation | <7.95 × 106 | −2.8 | Unlikely |
| HBB | Familial erythrocytosis | Pathogenic | Y145H | VANALAHKYH> | Phosphotyrosine | <7.95 × 106 | 4.68 | New hypothesis |
| HBB | Familial erythrocytosis | Pathogenic | Y145C | VANALAHKYH> | Phosphotyrosine | <7.95 × 106 | 4.68 | New hypothesis |
| HBB | Familial erythrocytosis | Pathogenic | Y145N | VANALAHKYH> | Phosphotyrosine | <7.95 × 106 | 4.68 | New hypothesis |
| INS | Permanent neonatal diabetes mellitus | Pathogenic | Y108C | SLYQLENYCN> | Phosphotyrosine | <7.95 × 106 | 3.58 | Unlikely |
| RHO | Retinitis pigmentosa | Likely pathogenic | S343N | SKTETSQVAPA> | Phosphoserine; dephosphorylation | <7.95 × 106 | 5.42 | Rediscovery |
| SDHA | Mitochondrial complex II deficiency; paragangliomas 5 | Likely pathogenic; uncertain significance | R662C | ATVPPAIRSY> | Methylation | <7.95 × 106 | 4.12 | New hypothesis |
| SMAD3 | Familial thoracic aneurysm and aortic dissection | Likely pathogenic | S423N | SPSIRCSSVS> | Phosphoserine | <7.95 × 106 | 4.97 | New hypothesis |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
FitzHugh, Z.T.; Schiller, M.R. Systematic Assessment of Protein C-Termini Mutated in Human Disorders. Biomolecules 2023, 13, 355. https://doi.org/10.3390/biom13020355
FitzHugh ZT, Schiller MR. Systematic Assessment of Protein C-Termini Mutated in Human Disorders. Biomolecules. 2023; 13(2):355. https://doi.org/10.3390/biom13020355
Chicago/Turabian StyleFitzHugh, Zachary T., and Martin R. Schiller. 2023. "Systematic Assessment of Protein C-Termini Mutated in Human Disorders" Biomolecules 13, no. 2: 355. https://doi.org/10.3390/biom13020355
APA StyleFitzHugh, Z. T., & Schiller, M. R. (2023). Systematic Assessment of Protein C-Termini Mutated in Human Disorders. Biomolecules, 13(2), 355. https://doi.org/10.3390/biom13020355