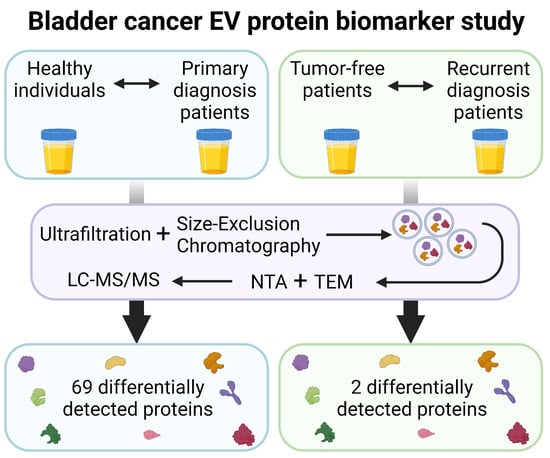
Protein Biomarker Discovery Studies on Urinary sEV Fractions Separated with UF-SEC for the First Diagnosis and Detection of Recurrence in Bladder Cancer Patients
Abstract
:1. Introduction
2. Materials and Methods
2.1. Urine Collection
2.2. Extracellular Vesicle (EV) Isolation
2.3. sEV Characterization
2.3.1. Protein Concentration
2.3.2. Transmission Electron Microscopy (TEM)
2.3.3. Nanoparticle Tracking Analysis (NTA)
2.4. Proteomic Analysis
2.4.1. Sample Preparation and Liquid Chromatography Followed by Tandem Mass Spectrometry (LC MS/MS)
2.4.2. Quality Controls
2.4.3. Peptide Identification
2.4.4. Data Enrichment for Peptide Quantification
2.5. Statistical Analysis
2.6. Network Analysis
2.7. EV-TRACK
3. Results
3.1. Demographics
3.2. sEV Characterization
3.3. Standard Deviation of Patient Samples
3.4. Statistically Differentially Expressed Proteins
3.5. Network Analysis
3.5.1. Networks and Pathways
3.5.2. Differentially Expressed Proteins Not in Network Analysis
3.6. Discriminating Power of Biomarker Candidates
4. Discussion
4.1. Standard Deviation of Patient Samples and Statistically Differentially Expressed Proteins
4.2. Network Analysis
4.2.1. Ubiquitination and ESCRT-Pathway-Related Proteins
4.2.2. Fibrinolysis, Complement Activation and Coagulation Cascades
4.2.3. Hemoglobins
4.2.4. Peptidases
4.2.5. Pro-Epidermal Growth Factor (EGF) and Prominin-1 (PROM1)
4.2.6. Differentially Expressed Proteins Not in Network Analysis
4.3. Potential Biomarkers
4.4. Future Research
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ANOVA | Analysis of variance |
| ATP | Adenosine triphosphate |
| BC | Bladder cancer |
| cf | Cell-free |
| EMT | Epithelial-to-mesenchymal transition |
| EV | Extracellular vesicle |
| FDR | False discovery rate |
| HC | Healthy control |
| HCD | Higher-energy collisional dissociation |
| LC-MS/MS | Liquid chromatography followed by tandem mass spectrometry |
| MCL | Markov clustering |
| MIBC | Muscle-invasive bladder cancer |
| MTBE | Methyl-tert-butyl-ether |
| MVBs | multivesicular bodies |
| NMIBC | Non-muscle-invasive bladder cancer |
| NTA | Nanoparticle tracking analysis |
| PD | Proteome discoverer |
| ppm | Parts per million |
| QC | Quality control |
| SEC | Size-exclusion chromatography |
| sEV | Small extracellular vesicles |
| STRING | Search tool for the retrieval of interacting genes |
| TEM | Transmission electron microscopy |
| UF | Ultrafiltration |
| UPS | Ubiquitin proteasome system |
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Global Burden of Disease Cancer, C.; Fitzmaurice, C.; Allen, C.; Barber, R.M.; Barregard, L.; Bhutta, Z.A.; Brenner, H.; Dicker, D.J.; Chimed-Orchir, O.; Dandona, R.; et al. Global, Regional, and National Cancer Incidence, Mortality, Years of Life Lost, Years Lived With Disability, and Disability-Adjusted Life-years for 32 Cancer Groups, 1990 to 2015: A Systematic Analysis for the Global Burden of Disease Study. JAMA Oncol. 2017, 3, 524–548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zuiverloon, T.C.; van der Aa, M.N.; van der Kwast, T.H.; Steyerberg, E.W.; Lingsma, H.F.; Bangma, C.H.; Zwarthoff, E.C. Fibroblast growth factor receptor 3 mutation analysis on voided urine for surveillance of patients with low-grade non-muscle-invasive bladder cancer. Clin. Cancer Res. 2010, 16, 3011–3018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ratert, N.; Meyer, H.A.; Jung, M.; Lioudmer, P.; Mollenkopf, H.J.; Wagner, I.; Miller, K.; Kilic, E.; Erbersdobler, A.; Weikert, S.; et al. miRNA profiling identifies candidate mirnas for bladder cancer diagnosis and clinical outcome. J. Mol. Diagn. 2013, 15, 695–705. [Google Scholar] [CrossRef] [PubMed]
- Babjuk, M.; Oosterlinck, W.; Sylvester, R.; Kaasinen, E.; Bohle, A.; Palou-Redorta, J.; Roupret, M. EAU guidelines on non-muscle-invasive urothelial carcinoma of the bladder, the 2011 update. Eur. Urol. 2011, 59, 997–1008. [Google Scholar] [CrossRef] [PubMed]
- Pollard, C.; Smith, S.C.; Theodorescu, D. Molecular genesis of non-muscle-invasive urothelial carcinoma (NMIUC). Expert. Rev. Mol. Med. 2010, 12, e10. [Google Scholar] [CrossRef] [Green Version]
- Baumgart, S.; Holters, S.; Ohlmann, C.H.; Bohle, R.; Stockle, M.; Ostenfeld, M.S.; Dyrskjot, L.; Junker, K.; Heinzelmann, J. Exosomes of invasive urothelial carcinoma cells are characterized by a specific miRNA expression signature. Oncotarget 2017, 8, 58278–58291. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, T.; Kates, M.; Fujita, K.; Bivalacqua, T.J.; McConkey, D.J. Predictive biomarkers for drug response in bladder cancer. Int. J. Urol. 2019, 26, 1044–1053. [Google Scholar] [CrossRef]
- Biardeau, X.; Lam, O.; Ba, V.; Campeau, L.; Corcos, J. Prospective evaluation of anxiety, pain, and embarrassment associated with cystoscopy and urodynamic testing in clinical practice. Can. Urol. Assoc. J. 2017, 11, 104–110. [Google Scholar] [CrossRef]
- Lotan, Y.; Roehrborn, C.G. Sensitivity and specificity of commonly available bladder tumor markers versus cytology: Results of a comprehensive literature review and meta-analyses. Urology 2003, 61, 109–118. [Google Scholar] [CrossRef]
- Oeyen, E.; Hoekx, L.; De Wachter, S.; Baldewijns, M.; Ameye, F.; Mertens, I. Bladder Cancer Diagnosis and Follow-Up: The Current Status and Possible Role of Extracellular Vesicles. Int. J. Mol. Sci. 2019, 20, 821. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lane, R.E.; Korbie, D.; Hill, M.M.; Trau, M. Extracellular vesicles as circulating cancer biomarkers: Opportunities and challenges. Clin. Transl. Med. 2018, 7, 14. [Google Scholar] [CrossRef] [PubMed]
- Bobrie, A.; Colombo, M.; Raposo, G.; Thery, C. Exosome secretion: Molecular mechanisms and roles in immune responses. Traffic 2011, 12, 1659–1668. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.R.; Ortiz-Bonilla, C.J.; Lee, Y.F. Extracellular Vesicles in Bladder Cancer: Biomarkers and Beyond. Int. J. Mol. Sci. 2018, 19, 2822. [Google Scholar] [CrossRef] [Green Version]
- Kalluri, R. The biology and function of exosomes in cancer. J. Clin. Invest. 2016, 126, 1208–1215. [Google Scholar] [CrossRef]
- Linxweiler, J.; Junker, K. Extracellular vesicles in urological malignancies: An update. Nat. Rev. Urol. 2020, 17, 11–27. [Google Scholar] [CrossRef]
- Callister, S.J.; Barry, R.C.; Adkins, J.N.; Johnson, E.T.; Qian, W.J.; Webb-Robertson, B.J.; Smith, R.D.; Lipton, M.S. Normalization approaches for removing systematic biases associated with mass spectrometry and label-free proteomics. J. Proteome Res. 2006, 5, 277–286. [Google Scholar] [CrossRef] [Green Version]
- Valikangas, T.; Suomi, T.; Elo, L.L. A systematic evaluation of normalization methods in quantitative label-free proteomics. Brief. Bioinform. 2018, 19, 1–11. [Google Scholar] [CrossRef]
- Tyanova, S.; Cox, J. Perseus: A Bioinformatics Platform for Integrative Analysis of Proteomics Data in Cancer Research. Methods Mol. Biol. 2018, 1711, 133–148. [Google Scholar] [CrossRef] [Green Version]
- Nawaz, M.; Camussi, G.; Valadi, H.; Nazarenko, I.; Ekstrom, K.; Wang, X.; Principe, S.; Shah, N.; Ashraf, N.M.; Fatima, F.; et al. The emerging role of extracellular vesicles as biomarkers for urogenital cancers. Nat. Rev. Urol. 2014, 11, 688–701. [Google Scholar] [CrossRef]
- Lee, J.; McKinney, K.Q.; Pavlopoulos, A.J.; Niu, M.; Kang, J.W.; Oh, J.W.; Kim, K.P.; Hwang, S. Altered Proteome of Extracellular Vesicles Derived from Bladder Cancer Patients Urine. Mol. Cells 2018, 41, 179–187. [Google Scholar] [CrossRef]
- Chen, C.L.; Lai, Y.F.; Tang, P.; Chien, K.Y.; Yu, J.S.; Tsai, C.H.; Chen, H.W.; Wu, C.C.; Chung, T.; Hsu, C.W.; et al. Comparative and targeted proteomic analyses of urinary microparticles from bladder cancer and hernia patients. J. Proteome Res. 2012, 11, 5611–5629. [Google Scholar] [CrossRef]
- Sun, Y.; Liu, J. Potential of cancer cell-derived exosomes in clinical application: A review of recent research advances. Clin. Ther. 2014, 36, 863–872. [Google Scholar] [CrossRef]
- Huebner, A.R.; Somparn, P.; Benjachat, T.; Leelahavanichkul, A.; Avihingsanon, Y.; Fenton, R.A.; Pisitkun, T. Exosomes in urine biomarker discovery. Adv. Exp. Med. Biol. 2015, 845, 43–58. [Google Scholar] [CrossRef]
- Franzen, C.A.; Blackwell, R.H.; Foreman, K.E.; Kuo, P.C.; Flanigan, R.C.; Gupta, G.N. Urinary Exosomes: The Potential for Biomarker Utility, Intercellular Signaling and Therapeutics in Urological Malignancy. J. Urol. 2016, 195, 1331–1339. [Google Scholar] [CrossRef]
- Hiltbrunner, S.; Mints, M.; Eldh, M.; Rosenblatt, R.; Holmstrom, B.; Alamdari, F.; Johansson, M.; Veerman, R.E.; Winqvist, O.; Sherif, A.; et al. Urinary Exosomes from Bladder Cancer Patients Show a Residual Cancer Phenotype despite Complete Pathological Downstaging. Sci. Rep. 2020, 10, 5960. [Google Scholar] [CrossRef] [Green Version]
- Tomiyama, E.; Matsuzaki, K.; Fujita, K.; Shiromizu, T.; Narumi, R.; Jingushi, K.; Koh, Y.; Matsushita, M.; Nakano, K.; Hayashi, Y.; et al. Proteomic analysis of urinary and tissue-exudative extracellular vesicles to discover novel bladder cancer biomarkers. Cancer Sci. 2021, 112, 2033–2045. [Google Scholar] [CrossRef]
- Tomiyama, E.; Fujita, K.; Matsuzaki, K.; Narumi, R.; Yamamoto, A.; Uemura, T.; Yamamichi, G.; Koh, Y.; Matsushita, M.; Hayashi, Y.; et al. EphA2 on urinary extracellular vesicles as a novel biomarker for bladder cancer diagnosis and its effect on the invasiveness of bladder cancer. Br. J. Cancer 2022, 127, 1312–1323. [Google Scholar] [CrossRef]
- Lee, J.; Park, H.S.; Han, S.R.; Kang, Y.H.; Mun, J.Y.; Shin, D.W.; Oh, H.W.; Cho, Y.K.; Lee, M.S.; Park, J. Alpha-2-macroglobulin as a novel diagnostic biomarker for human bladder cancer in urinary extracellular vesicles. Front. Oncol. 2022, 12, 976407. [Google Scholar] [CrossRef]
- Thery, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef] [Green Version]
- Goethals, S.; De Wilde, A.; Lesage, K.; Smits, E.; Pauwels, P.; Peeters, M. Tumorbank@uza: A Collection of Tissue, Fluid Samples and Associated Data of Oncology Patients for the Use in Translational Research. Open. J. Bioresour. 2018, 5, 4. [Google Scholar] [CrossRef] [Green Version]
- Yuana, Y.; Boing, A.N.; Grootemaat, A.E.; van der Pol, E.; Hau, C.M.; Cizmar, P.; Buhr, E.; Sturk, A.; Nieuwland, R. Handling and storage of human body fluids for analysis of extracellular vesicles. J. Extracell. Vesicles 2015, 4, 29260. [Google Scholar] [CrossRef] [PubMed]
- Webber, J.; Clayton, A. How pure are your vesicles? J. Extracell. Vesicles 2013, 2, 19861. [Google Scholar] [CrossRef]
- Oeyen, E.; Van Mol, K.; Baggerman, G.; Willems, H.; Boonen, K.; Rolfo, C.; Pauwels, P.; Jacobs, A.; Schildermans, K.; Cho, W.C.; et al. Ultrafiltration and size exclusion chromatography combined with asymmetrical-flow field-flow fractionation for the isolation and characterisation of extracellular vesicles from urine. J. Extracell. Vesicles 2018, 7, 1490143. [Google Scholar] [CrossRef] [PubMed]
- Oeyen, E.; Willems, H.; Kindt, R.T.; Sandra, K.; Boonen, K.; Hoekx, L.; De Wachter, S.; Ameye, F.; Mertens, I. Determination of variability due to biological and technical variation in urinary extracellular vesicles as a crucial step in biomarker discovery studies. J. Extracell. Vesicles 2019, 8, 1676035. [Google Scholar] [CrossRef] [Green Version]
- Mertens, I.; Willems, H.; Van Loon, E.; Schildermans, K.; Boonen, K.; Baggerman, G.; Valkenborg, D.; Gwinner, W.; Anglicheau, D.; Essig, M.; et al. Urinary Protein Biomarker Panel for the Diagnosis of Antibody-Mediated Rejection in Kidney Transplant Recipients. Kidney Int. Rep. 2020, 5, 1448–1458. [Google Scholar] [CrossRef]
- Gentleman, R.C.; Carey, V.J.; Bates, D.M.; Bolstad, B.; Dettling, M.; Dudoit, S.; Ellis, B.; Gautier, L.; Ge, Y.; Gentry, J.; et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004, 5, R80. [Google Scholar] [CrossRef] [Green Version]
- R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing 2018. Available online: https://www.R-project.org/ (accessed on 22 November 2022).
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017, 45, D362–D368. [Google Scholar] [CrossRef] [Green Version]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [Green Version]
- Consortium, E.-T.; Van Deun, J.; Mestdagh, P.; Agostinis, P.; Akay, O.; Anand, S.; Anckaert, J.; Martinez, Z.A.; Baetens, T.; Beghein, E.; et al. EV-TRACK: Transparent reporting and centralizing knowledge in extracellular vesicle research. Nat. Methods 2017, 14, 228–232. [Google Scholar] [CrossRef]
- Boulestreau, J.; Maumus, M.; Rozier, P.; Jorgensen, C.; Noel, D. Mesenchymal Stem Cell Derived Extracellular Vesicles in Aging. Front. Cell Dev. Biol. 2020, 8, 107. [Google Scholar] [CrossRef] [Green Version]
- Gallo, L.H.; Ko, J.; Donoghue, D.J. The importance of regulatory ubiquitination in cancer and metastasis. Cell Cycle 2017, 16, 634–648. [Google Scholar] [CrossRef] [Green Version]
- Sharma, A.; Trivedi, A.K. Regulation of apoptosis by E3 ubiquitin ligases in ubiquitin proteasome system. Cell Biol. Int. 2020, 44, 721–734. [Google Scholar] [CrossRef]
- Ichioka, F.; Kobayashi, R.; Katoh, K.; Shibata, H.; Maki, M. Brox, a novel farnesylated Bro1 domain-containing protein that associates with charged multivesicular body protein 4 (CHMP4). FEBS J. 2008, 275, 682–692. [Google Scholar] [CrossRef]
- Broniarczyk, J.; Olejnik-Schmidt, A.K.; Luczak, M.W.; Schmidt, M.T.; Dabrowski, M.; Jozefiak, A.; Kedzia, W.; Kwasniewska, A.; Gozdzicka-Jozefiak, A. Analysis of expression and structure of the TSG101 gene in cervical cancer cells. Int. J. Mol. Med. 2010, 25, 777–783. [Google Scholar] [CrossRef]
- Jiang, Y.; Ou, Y.; Cheng, X. Role of TSG101 in cancer. Front. Biosci. 2013, 18, 279–288. [Google Scholar] [CrossRef] [Green Version]
- Latosinska, A.; Mokou, M.; Makridakis, M.; Mullen, W.; Zoidakis, J.; Lygirou, V.; Frantzi, M.; Katafigiotis, I.; Stravodimos, K.; Hupe, M.C.; et al. Proteomics analysis of bladder cancer invasion: Targeting EIF3D for therapeutic intervention. Oncotarget 2017, 8, 69435–69455. [Google Scholar] [CrossRef] [Green Version]
- Colombo, M.; Moita, C.; van Niel, G.; Kowal, J.; Vigneron, J.; Benaroch, P.; Manel, N.; Moita, L.F.; Thery, C.; Raposo, G. Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles. J. Cell. Sci. 2013, 126, 5553–5565. [Google Scholar] [CrossRef] [Green Version]
- Jeong, D.H.; Kim, H.K.; Prince, A.E.; Lee, D.S.; Kim, Y.N.; Han, J.; Kim, K.T. Plasma proteomic analysis of patients with squamous cell carcinoma of the uterine cervix. J. Gynecol. Oncol. 2008, 19, 173–180. [Google Scholar] [CrossRef] [Green Version]
- Bergamini, S.; Bellei, E.; Reggiani Bonetti, L.; Monari, E.; Cuoghi, A.; Borelli, F.; Sighinolfi, M.C.; Bianchi, G.; Ozben, T.; Tomasi, A. Inflammation: An important parameter in the search of prostate cancer biomarkers. Proteome Sci. 2014, 12, 32. [Google Scholar] [CrossRef] [Green Version]
- Davalieva, K.; Kiprijanovska, S.; Maleva Kostovska, I.; Stavridis, S.; Stankov, O.; Komina, S.; Petrusevska, G.; Polenakovic, M. Comparative Proteomics Analysis of Urine Reveals Down-Regulation of Acute Phase Response Signaling and LXR/RXR Activation Pathways in Prostate Cancer. Proteomes 2017, 6, 1. [Google Scholar] [CrossRef] [Green Version]
- Dowling, P.; Clarke, C.; Hennessy, K.; Torralbo-Lopez, B.; Ballot, J.; Crown, J.; Kiernan, I.; O’Byrne, K.J.; Kennedy, M.J.; Lynch, V.; et al. Analysis of acute-phase proteins, AHSG, C3, CLI, HP and SAA, reveals distinctive expression patterns associated with breast, colorectal and lung cancer. Int. J. Cancer 2012, 131, 911–923. [Google Scholar] [CrossRef] [Green Version]
- Kim, P.Y.; Tan, O.; Diakiw, S.M.; Carter, D.; Sekerye, E.O.; Wasinger, V.C.; Liu, T.; Kavallaris, M.; Norris, M.D.; Haber, M.; et al. Identification of plasma complement C3 as a potential biomarker for neuroblastoma using a quantitative proteomic approach. J. Proteomics 2014, 96, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Taylor, R.P.; Lindorfer, M.A. Cytotoxic mechanisms of immunotherapy: Harnessing complement in the action of anti-tumor monoclonal antibodies. Semin. Immunol. 2016, 28, 309–316. [Google Scholar] [CrossRef]
- Corrales, L.; Ajona, D.; Rafail, S.; Lasarte, J.J.; Riezu-Boj, J.I.; Lambris, J.D.; Rouzaut, A.; Pajares, M.J.; Montuenga, L.M.; Pio, R. Anaphylatoxin C5a creates a favorable microenvironment for lung cancer progression. J. Immunol. 2012, 189, 4674–4683. [Google Scholar] [CrossRef] [Green Version]
- Grivennikov, S.I.; Greten, F.R.; Karin, M. Immunity, inflammation, and cancer. Cell 2010, 140, 883–899. [Google Scholar] [CrossRef] [Green Version]
- Song, Y.H.; Shiota, M.; Yokomizo, A.; Uchiumi, T.; Kiyoshima, K.; Kuroiwa, K.; Oda, Y.; Naito, S. Twist1 and Y-box-binding protein-1 are potential prognostic factors in bladder cancer. Urol. Oncol. 2014, 32, 31.e1–31.e7. [Google Scholar] [CrossRef]
- van Kessel, K.E.; Van Neste, L.; Lurkin, I.; Zwarthoff, E.C.; Van Criekinge, W. Evaluation of an Epigenetic Profile for the Detection of Bladder Cancer in Patients with Hematuria. J. Urol. 2016, 195, 601–607. [Google Scholar] [CrossRef]
- Berx, G.; Cleton-Jansen, A.M.; Nollet, F.; de Leeuw, W.J.; van de Vijver, M.; Cornelisse, C.; van Roy, F. E-cadherin is a tumour/invasion suppressor gene mutated in human lobular breast cancers. EMBO J. 1995, 14, 6107–6115. [Google Scholar] [CrossRef]
- Cho, M.S.; Rupaimoole, R.; Choi, H.J.; Noh, K.; Chen, J.; Hu, Q.; Sood, A.K.; Afshar-Kharghan, V. Complement Component 3 Is Regulated by TWIST1 and Mediates Epithelial-Mesenchymal Transition. J. Immunol. 2016, 196, 1412–1418. [Google Scholar] [CrossRef] [Green Version]
- Fishelson, Z.; Donin, N.; Zell, S.; Schultz, S.; Kirschfink, M. Obstacles to cancer immunotherapy: Expression of membrane complement regulatory proteins (mCRPs) in tumors. Mol. Immunol. 2003, 40, 109–123. [Google Scholar] [CrossRef]
- Madjd, Z.; Pinder, S.E.; Paish, C.; Ellis, I.O.; Carmichael, J.; Durrant, L.G. Loss of CD59 expression in breast tumours correlates with poor survival. J. Pathol. 2003, 200, 633–639. [Google Scholar] [CrossRef]
- Abdullah-Soheimi, S.S.; Lim, B.K.; Hashim, O.H.; Shuib, A.S. Patients with ovarian carcinoma excrete different altered levels of urine CD59, kininogen-1 and fragments of inter-alpha-trypsin inhibitor heavy chain H4 and albumin. Proteome Sci. 2010, 8, 58. [Google Scholar] [CrossRef] [Green Version]
- Geller, A.; Yan, J. The Role of Membrane Bound Complement Regulatory Proteins in Tumor Development and Cancer Immunotherapy. Front. Immunol. 2019, 10, 1074. [Google Scholar] [CrossRef] [Green Version]
- Davalieva, K.; Kiprijanovska, S.; Komina, S.; Petrusevska, G.; Zografska, N.C.; Polenakovic, M. Proteomics analysis of urine reveals acute phase response proteins as candidate diagnostic biomarkers for prostate cancer. Proteome Sci. 2015, 13, 2. [Google Scholar] [CrossRef] [Green Version]
- Ruivenkamp, C.A.; van Wezel, T.; Zanon, C.; Stassen, A.P.; Vlcek, C.; Csikos, T.; Klous, A.M.; Tripodis, N.; Perrakis, A.; Boerrigter, L.; et al. Ptprj is a candidate for the mouse colon-cancer susceptibility locus Scc1 and is frequently deleted in human cancers. Nat. Genet. 2002, 31, 295–300. [Google Scholar] [CrossRef]
- Kwaan, H.C.; Lindholm, P.F. Fibrin and Fibrinolysis in Cancer. Semin. Thromb. Hemost. 2019, 45, 413–422. [Google Scholar] [CrossRef]
- Repetto, O.; De Re, V. Coagulation and fibrinolysis in gastric cancer. Ann. N. Y Acad. Sci. 2017, 1404, 27–48. [Google Scholar] [CrossRef]
- Guglietta, S.; Rescigno, M. Hypercoagulation and complement: Connected players in tumor development and metastases. Semin. Immunol. 2016, 28, 578–586. [Google Scholar] [CrossRef]
- Steinbrecher, K.A.; Horowitz, N.A.; Blevins, E.A.; Barney, K.A.; Shaw, M.A.; Harmel-Laws, E.; Finkelman, F.D.; Flick, M.J.; Pinkerton, M.D.; Talmage, K.E.; et al. Colitis-associated cancer is dependent on the interplay between the hemostatic and inflammatory systems and supported by integrin alpha(M)beta(2) engagement of fibrinogen. Cancer Res. 2010, 70, 2634–2643. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Li, D.; Cao, L.; Wang, Z.; Li, Y.; Liu, H.; Chen, G. Elevated preoperative plasma fibrinogen level is an independent predictor of malignancy and advanced stage disease in patients with bladder urothelial tumors. Int. J. Surg. 2016, 36, 249–254. [Google Scholar] [CrossRef]
- Li, H.; Li, C.; Wu, H.; Zhang, T.; Wang, J.; Wang, S.; Chang, J. Identification of Apo-A1 as a biomarker for early diagnosis of bladder transitional cell carcinoma. Proteome Sci. 2011, 9, 21. [Google Scholar] [CrossRef] [Green Version]
- Odabaei, G.; Chatterjee, D.; Jazirehi, A.R.; Goodglick, L.; Yeung, K.; Bonavida, B. Raf-1 kinase inhibitor protein: Structure, function, regulation of cell signaling, and pivotal role in apoptosis. Adv. Cancer Res. 2004, 91, 169–200. [Google Scholar] [CrossRef]
- Yeung, K.; Seitz, T.; Li, S.; Janosch, P.; McFerran, B.; Kaiser, C.; Fee, F.; Katsanakis, K.D.; Rose, D.W.; Mischak, H.; et al. Suppression of Raf-1 kinase activity and MAP kinase signalling by RKIP. Nature 1999, 401, 173–177. [Google Scholar] [CrossRef] [Green Version]
- Park, S.; Yeung, M.L.; Beach, S.; Shields, J.M.; Yeung, K.C. RKIP downregulates B-Raf kinase activity in melanoma cancer cells. Oncogene 2005, 24, 3535–3540. [Google Scholar] [CrossRef] [Green Version]
- Lamiman, K.; Keller, J.M.; Mizokami, A.; Zhang, J.; Keller, E.T. Survey of Raf kinase inhibitor protein (RKIP) in multiple cancer types. Crit. Rev. Oncog. 2014, 19, 455–468. [Google Scholar] [CrossRef]
- Li, H.Z.; Wang, Y.; Gao, Y.; Shao, J.; Zhao, X.L.; Deng, W.M.; Liu, Y.X.; Yang, J.; Yao, Z. Effects of raf kinase inhibitor protein expression on metastasis and progression of human epithelial ovarian cancer. Mol. Cancer Res. 2008, 6, 917–928. [Google Scholar] [CrossRef] [Green Version]
- Zaravinos, A.; Chatziioannou, M.; Lambrou, G.I.; Boulalas, I.; Delakas, D.; Spandidos, D.A. Implication of RAF and RKIP genes in urinary bladder cancer. Pathol. Oncol. Res. 2011, 17, 181–190. [Google Scholar] [CrossRef]
- Al-Mulla, F.; Bitar, M.S.; Taqi, Z.; Yeung, K.C. RKIP: Much more than Raf kinase inhibitory protein. J. Cell Physiol. 2013, 228, 1688–1702. [Google Scholar] [CrossRef]
- Farooqi, A.A.; Li, Y.; Sarkar, F.H. The biological complexity of RKIP signaling in human cancers. Exp. Mol. Med. 2015, 47, e185. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.C.; Qi, X.; Juarez, J.; Plunkett, M.; Donate, F.; Sakthivel, R.; Mazar, A.P.; McCrae, K.R. Inhibition of angiogenesis by two-chain high molecular weight kininogen (HKa) and kininogen-derived polypeptides. Can. J. Physiol. Pharmacol. 2002, 80, 85–90. [Google Scholar] [CrossRef]
- Roeise, O.; Sivertsen, S.; Ruud, T.E.; Bouma, B.N.; Stadaas, J.O.; Aasen, A.O. Studies on components of the contact phase system in patients with advanced gastrointestinal cancer. Cancer 1990, 65, 1355–1359. [Google Scholar] [CrossRef]
- Doustjalali, S.R.; Yusof, R.; Yip, C.H.; Looi, L.M.; Pillay, B.; Hashim, O.H. Aberrant expression of acute-phase reactant proteins in sera and breast lesions of patients with malignant and benign breast tumors. Electrophoresis 2004, 25, 2392–2401. [Google Scholar] [CrossRef]
- Abdul-Rahman, P.S.; Lim, B.K.; Hashim, O.H. Expression of high-abundance proteins in sera of patients with endometrial and cervical cancers: Analysis using 2-DE with silver staining and lectin detection methods. Electrophoresis 2007, 28, 1989–1996. [Google Scholar] [CrossRef] [Green Version]
- Weeks, M.E.; Hariharan, D.; Petronijevic, L.; Radon, T.P.; Whiteman, H.J.; Kocher, H.M.; Timms, J.F.; Lemoine, N.R.; Crnogorac-Jurcevic, T. Analysis of the urine proteome in patients with pancreatic ductal adenocarcinoma. Proteomics Clin. Appl. 2008, 2, 1047–1057. [Google Scholar] [CrossRef]
- Canter, M.P.; Graham, C.A.; Heit, M.H.; Blackwell, L.S.; Wilkey, D.W.; Klein, J.B.; Merchant, M.L. Proteomic techniques identify urine proteins that differentiate patients with interstitial cystitis from asymptomatic control subjects. Am. J. Obstet. Gynecol. 2008, 198, 553.E1–553.E6. [Google Scholar] [CrossRef]
- Rocchetti, M.T.; Centra, M.; Papale, M.; Bortone, G.; Palermo, C.; Centonze, D.; Ranieri, E.; Di Paolo, S.; Gesualdo, L. Urine protein profile of IgA nephropathy patients may predict the response to ACE-inhibitor therapy. Proteomics 2008, 8, 206–216. [Google Scholar] [CrossRef]
- Chen, C.L.; Lin, T.S.; Tsai, C.H.; Wu, C.C.; Chung, T.; Chien, K.Y.; Wu, M.; Chang, Y.S.; Yu, J.S.; Chen, Y.T. Identification of potential bladder cancer markers in urine by abundant-protein depletion coupled with quantitative proteomics. J. Proteomics 2013, 85, 28–43. [Google Scholar] [CrossRef] [Green Version]
- Russo, J.W.; Gao, C.; Bhasin, S.S.; Voznesensky, O.S.; Calagua, C.; Arai, S.; Nelson, P.S.; Montgomery, B.; Mostaghel, E.A.; Corey, E.; et al. Downregulation of Dipeptidyl Peptidase 4 Accelerates Progression to Castration-Resistant Prostate Cancer. Cancer Res. 2018, 78, 6354–6362. [Google Scholar] [CrossRef] [Green Version]
- Masur, K.; Schwartz, F.; Entschladen, F.; Niggemann, B.; Zaenker, K.S. DPPIV inhibitors extend GLP-2 mediated tumour promoting effects on intestinal cancer cells. Regul. Pept. 2006, 137, 147–155. [Google Scholar] [CrossRef]
- Havre, P.A.; Abe, M.; Urasaki, Y.; Ohnuma, K.; Morimoto, C.; Dang, N.H. The role of CD26/dipeptidyl peptidase IV in cancer. Front. Biosci. 2008, 13, 1634–1645. [Google Scholar] [CrossRef]
- Ohnuma, K.; Takahashi, N.; Yamochi, T.; Hosono, O.; Dang, N.H.; Morimoto, C. Role of CD26/dipeptidyl peptidase IV in human T cell activation and function. Front. Biosci. 2008, 13, 2299–2310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sorensen, K.D.; Abildgaard, M.O.; Haldrup, C.; Ulhoi, B.P.; Kristensen, H.; Strand, S.; Parker, C.; Hoyer, S.; Borre, M.; Orntoft, T.F. Prognostic significance of aberrantly silenced ANPEP expression in prostate cancer. Br. J. Cancer 2013, 108, 420–428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Z.; Yang, Z.; Xiong, L.; Li, D.; Zou, Q.; Yuan, Y. ACO2 and ANPEP as novel prognostic markers for gallbladder squamous cell/adenosquamous carcinomas and adenocarcinomas. Int. J. Clin. Oncol. 2020, 25, 1346–1355. [Google Scholar] [CrossRef]
- Freedland, S.J.; Seligson, D.B.; Liu, A.Y.; Pantuck, A.J.; Paik, S.H.; Horvath, S.; Wieder, J.A.; Zisman, A.; Nguyen, D.; Tso, C.L.; et al. Loss of CD10 (neutral endopeptidase) is a frequent and early event in human prostate cancer. Prostate 2003, 55, 71–80. [Google Scholar] [CrossRef]
- Gohring, B.; Holzhausen, H.J.; Meye, A.; Heynemann, H.; Rebmann, U.; Langner, J.; Riemann, D. Endopeptidase 24.11/CD10 is down-regulated in renal cell cancer. Int. J. Mol. Med. 1998, 2, 409–414. [Google Scholar] [CrossRef]
- Papandreou, C.N.; Usmani, B.; Geng, Y.; Bogenrieder, T.; Freeman, R.; Wilk, S.; Finstad, C.L.; Reuter, V.E.; Powell, C.T.; Scheinberg, D.; et al. Neutral endopeptidase 24.11 loss in metastatic human prostate cancer contributes to androgen-independent progression. Nat. Med. 1998, 4, 50–57. [Google Scholar] [CrossRef]
- Bilalovic, N.; Sandstad, B.; Golouh, R.; Nesland, J.M.; Selak, I.; Torlakovic, E.E. CD10 protein expression in tumor and stromal cells of malignant melanoma is associated with tumor progression. Mod. Pathol. 2004, 17, 1251–1258. [Google Scholar] [CrossRef] [Green Version]
- Tomoda, C.; Kushima, R.; Takeuti, E.; Mukaisho, K.; Hattori, T.; Kitano, H. CD10 expression is useful in the diagnosis of follicular carcinoma and follicular variant of papillary thyroid carcinoma. Thyroid. 2003, 13, 291–295. [Google Scholar] [CrossRef]
- Lu, S.C. Regulation of glutathione synthesis. Mol. Aspects Med. 2009, 30, 42–59. [Google Scholar] [CrossRef] [Green Version]
- Smith, L.M.; Nesterova, A.; Ryan, M.C.; Duniho, S.; Jonas, M.; Anderson, M.; Zabinski, R.F.; Sutherland, M.K.; Gerber, H.P.; Van Orden, K.L.; et al. CD133/prominin-1 is a potential therapeutic target for antibody-drug conjugates in hepatocellular and gastric cancers. Br. J. Cancer 2008, 99, 100–109. [Google Scholar] [CrossRef] [Green Version]
- Saha, S.K.; Islam, S.M.R.; Kwak, K.S.; Rahman, M.S.; Cho, S.G. PROM1 and PROM2 expression differentially modulates clinical prognosis of cancer: A multiomics analysis. Cancer Gene Ther. 2020, 27, 147–167. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.T.; Chen, C.L.; Chen, H.W.; Chung, T.; Wu, C.C.; Chen, C.D.; Hsu, C.W.; Chen, M.C.; Tsui, K.H.; Chang, P.L.; et al. Discovery of novel bladder cancer biomarkers by comparative urine proteomics using iTRAQ technology. J. Proteome Res. 2010, 9, 5803–5815. [Google Scholar] [CrossRef]
- Sun, C.; Song, C.; Ma, Z.; Xu, K.; Zhang, Y.; Jin, H.; Tong, S.; Ding, W.; Xia, G.; Ding, Q. Periostin identified as a potential biomarker of prostate cancer by iTRAQ-proteomics analysis of prostate biopsy. Proteome Sci. 2011, 9, 22. [Google Scholar] [CrossRef] [Green Version]
- Yang, H.Y.; Kwon, J.; Park, H.R.; Kwon, S.O.; Park, Y.K.; Kim, H.S.; Chung, Y.J.; Chang, Y.J.; Choi, H.I.; Chung, K.J.; et al. Comparative proteomic analysis for the insoluble fractions of colorectal cancer patients. J. Proteomics 2012, 75, 3639–3653. [Google Scholar] [CrossRef]
- Parker, L.P.; Taylor, D.D.; Kesterson, S.; Gercel-Taylor, C. Gene expression profiling in response to estradiol and genistein in ovarian cancer cells. Cancer Genom. Proteom. 2009, 6, 189–194. [Google Scholar]
- He, J.; Jin, Y.; Chen, Y.; Yao, H.B.; Xia, Y.J.; Ma, Y.Y.; Wang, W.; Shao, Q.S. Downregulation of ALDOB is associated with poor prognosis of patients with gastric cancer. Onco Targets Ther. 2016, 9, 6099–6109. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.; Li, Y.; Xu, J.; Wang, S.; Xu, Y.; Li, X.; Cai, S. Aldolase B Overexpression is Associated with Poor Prognosis and Promotes Tumor Progression by Epithelial-Mesenchymal Transition in Colorectal Adenocarcinoma. Cell Physiol. Biochem. 2017, 42, 397–406. [Google Scholar] [CrossRef]
- God, J.M.; Cameron, C.; Figueroa, J.; Amria, S.; Hossain, A.; Kempkes, B.; Bornkamm, G.W.; Stuart, R.K.; Blum, J.S.; Haque, A. Elevation of c-MYC disrupts HLA class II-mediated immune recognition of human B cell tumors. J. Immunol. 2015, 194, 1434–1445. [Google Scholar] [CrossRef] [Green Version]
- Filipazzi, P.; Burdek, M.; Villa, A.; Rivoltini, L.; Huber, V. Recent advances on the role of tumor exosomes in immunosuppression and disease progression. Semin. Cancer Biol. 2012, 22, 342–349. [Google Scholar] [CrossRef] [Green Version]
- Meissner, M.; Whiteside, T.L.; Kaufmann, R.; Seliger, B. CIITA versus IFN-gamma induced MHC class II expression in head and neck cancer cells. Arch. Dermatol. Res. 2009, 301, 189–193. [Google Scholar] [CrossRef] [PubMed]
- Georgescu, M.M.; Gagea, M.; Cote, G. NHERF1/EBP50 Suppresses Wnt-beta-Catenin Pathway-Driven Intestinal Neoplasia. Neoplasia 2016, 18, 512–523. [Google Scholar] [CrossRef] [Green Version]
- Molina, J.R.; Agarwal, N.K.; Morales, F.C.; Hayashi, Y.; Aldape, K.D.; Cote, G.; Georgescu, M.M. PTEN, NHERF1 and PHLPP form a tumor suppressor network that is disabled in glioblastoma. Oncogene 2012, 31, 1264–1274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hayashi, Y.; Molina, J.R.; Hamilton, S.R.; Georgescu, M.M. NHERF1/EBP50 is a new marker in colorectal cancer. Neoplasia 2010, 12, 1013–1022. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tao, T.; Yang, X.; Qin, Q.; Shi, W.; Wang, Q.; Yang, Y.; He, J. NHERF1 Enhances Cisplatin Sensitivity in Human Cervical Cancer Cells. Int. J. Mol. Sci. 2017, 18, 5. [Google Scholar] [CrossRef] [Green Version]
- Prunotto, M.; Farina, A.; Lane, L.; Pernin, A.; Schifferli, J.; Hochstrasser, D.F.; Lescuyer, P.; Moll, S. Proteomic analysis of podocyte exosome-enriched fraction from normal human urine. J. Proteomics 2013, 82, 193–229. [Google Scholar] [CrossRef]
- Hwang, H.; Im, J.E.; Yang, Y.; Kim, H.; Kwon, K.H.; Kim, Y.H.; Kim, J.Y.; Yoo, J.S. Bioinformatic Prediction of Gene Ontology Terms of Uncharacterized Proteins from Chromosome 11. J. Proteome Res. 2020, 19, 4907–4912. [Google Scholar] [CrossRef]
- Gupta, R.; Radicioni, G.; Abdelwahab, S.; Dang, H.; Carpenter, J.; Chua, M.; Mieczkowski, P.A.; Sheridan, J.T.; Randell, S.H.; Kesimer, M. Intercellular Communication between Airway Epithelial Cells Is Mediated by Exosome-Like Vesicles. Am. J. Respir. Cell Mol. Biol. 2019, 60, 209–220. [Google Scholar] [CrossRef]
- Dhondt, B.; Geeurickx, E.; Tulkens, J.; Van Deun, J.; Vergauwen, G.; Lippens, L.; Miinalainen, I.; Rappu, P.; Heino, J.; Ost, P.; et al. Unravelling the proteomic landscape of extracellular vesicles in prostate cancer by density-based fractionation of urine. J. Extracell. Vesicles 2020, 9, 1736935. [Google Scholar] [CrossRef]
- Lv, L.H.; Wan, Y.L.; Lin, Y.; Zhang, W.; Yang, M.; Li, G.L.; Lin, H.M.; Shang, C.Z.; Chen, Y.J.; Min, J. Anticancer drugs cause release of exosomes with heat shock proteins from human hepatocellular carcinoma cells that elicit effective natural killer cell antitumor responses in vitro. J. Biol. Chem. 2012, 287, 15874–15885. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, K.; Yan, I.K.; Wood, J.; Haga, H.; Patel, T. Involvement of extracellular vesicle long noncoding RNA (linc-VLDLR) in tumor cell responses to chemotherapy. Mol. Cancer Res. 2014, 12, 1377–1387. [Google Scholar] [CrossRef] [Green Version]
- Kreger, B.T.; Johansen, E.R.; Cerione, R.A.; Antonyak, M.A. The Enrichment of Survivin in Exosomes from Breast Cancer Cells Treated with Paclitaxel Promotes Cell Survival and Chemoresistance. Cancers 2016, 8, 111. [Google Scholar] [CrossRef] [Green Version]
- Corcoran, C.; Rani, S.; O’Brien, K.; O’Neill, A.; Prencipe, M.; Sheikh, R.; Webb, G.; McDermott, R.; Watson, W.; Crown, J.; et al. Docetaxel-resistance in prostate cancer: Evaluating associated phenotypic changes and potential for resistance transfer via exosomes. PLoS ONE 2012, 7, e50999. [Google Scholar] [CrossRef] [Green Version]
- Ning, K.; Wang, T.; Sun, X.; Zhang, P.; Chen, Y.; Jin, J.; Hua, D. UCH-L1-containing exosomes mediate chemotherapeutic resistance transfer in breast cancer. J. Surg. Oncol. 2017, 115, 932–940. [Google Scholar] [CrossRef]
- Crow, J.; Atay, S.; Banskota, S.; Artale, B.; Schmitt, S.; Godwin, A.K. Exosomes as mediators of platinum resistance in ovarian cancer. Oncotarget 2017, 8, 11917–11936. [Google Scholar] [CrossRef] [Green Version]
- Cesi, G.; Philippidou, D.; Kozar, I.; Kim, Y.J.; Bernardin, F.; Van Niel, G.; Wienecke-Baldacchino, A.; Felten, P.; Letellier, E.; Dengler, S.; et al. A new ALK isoform transported by extracellular vesicles confers drug resistance to melanoma cells. Mol. Cancer 2018, 17, 145. [Google Scholar] [CrossRef]
- Wang, X.; Xu, C.; Hua, Y.; Sun, L.; Cheng, K.; Jia, Z.; Han, Y.; Dong, J.; Cui, Y.; Yang, Z. Exosomes play an important role in the process of psoralen reverse multidrug resistance of breast cancer. J. Exp. Clin. Cancer Res. 2016, 35, 186. [Google Scholar] [CrossRef] [Green Version]
- Kumar, P.; Nandi, S.; Tan, T.Z.; Ler, S.G.; Chia, K.S.; Lim, W.Y.; Butow, Z.; Vordos, D.; De la Taille, A.; Al-Haddawi, M.; et al. Highly sensitive and specific novel biomarkers for the diagnosis of transitional bladder carcinoma. Oncotarget 2015, 6, 13539–13549. [Google Scholar] [CrossRef] [Green Version]
- Habermann, J.K.; Roblick, U.J.; Luke, B.T.; Prieto, D.A.; Finlay, W.J.; Podust, V.N.; Roman, J.M.; Oevermann, E.; Schiedeck, T.; Homann, N.; et al. Increased serum levels of complement C3a anaphylatoxin indicate the presence of colorectal tumors. Gastroenterology 2006, 131, 1020–1029. [Google Scholar] [CrossRef] [Green Version]
- Villanueva, J.; Shaffer, D.R.; Philip, J.; Chaparro, C.A.; Erdjument-Bromage, H.; Olshen, A.B.; Fleisher, M.; Lilja, H.; Brogi, E.; Boyd, J.; et al. Differential exoprotease activities confer tumor-specific serum peptidome patterns. J. Clin. Invest. 2006, 116, 271–284. [Google Scholar] [CrossRef]
- Sallam, R.M. Proteomics in cancer biomarkers discovery: Challenges and applications. Dis. Markers 2015, 2015, 321370. [Google Scholar] [CrossRef] [PubMed]
- Lorentzian, A.; Uzozie, A.; Lange, P.F. Origins and clinical relevance of proteoforms in pediatric malignancies. Expert. Rev. Proteom. 2019, 16, 185–200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matthiesen, R.; Bunkenborg, J. Introduction to mass spectrometry-based proteomics. Methods Mol. Biol. 2013, 1007, 1–45. [Google Scholar] [CrossRef] [PubMed]
- Bringans, S.; Peters, K.; Casey, T.; Ito, J.; Lipscombe, R. The New and the Old: Platform Cross-Validation of Immunoaffinity MASS Spectrometry versus ELISA for PromarkerD, a Predictive Test for Diabetic Kidney Disease. Proteomes 2020, 8, 31. [Google Scholar] [CrossRef]
- Boys, E.L.; Liu, J.; Robinson, P.J.; Reddel, R.R. Clinical applications of mass spectrometry-based proteomics in cancer: Where are we? Proteomics 2023, 23, e2200238. [Google Scholar] [CrossRef]




| Healthy Controls | First Diagnosis of Bladder Cancer | Diagnosis of Bladder Cancer Relapse | Tumor Free | |
|---|---|---|---|---|
| X number of samples | 64 | 50 | 46 | 109 |
| Derived from X number of patients | 64 | 50 | 34 | 52 |
| Age (years) | ||||
| Average | 69.2 | 71.9 | 73.0 | 70.7 |
| Minimum | 40 | 42 | 48 | 35 |
| Maximum | 83 | 89 | 91 | 89 |
| Sex N (%) | ||||
| Male | 39 (61%) | 39 (78%) | 29 (85%) | 37 (71%) |
| Female | 25 (39%) | 11 (22%) | 5 (15%) | 15 (29%) |
| Tumor stage * | ||||
| Ta | NA | 15 | 25 | NA |
| T1 (including Tis) | NA | 30 | 19 | NA |
| T2 | NA | 5 | 2 | NA |
| Tumor grade * | ||||
| 1 | NA | 1 | 18 | NA |
| 2 | NA | 24 | 8 | NA |
| 3 | NA | 25 | 20 | NA |
| Hospital | ||||
| UZA | 32 | 5 | 25 | 17 |
| AZMM | 0 | 39 | 9 | 34 |
| AZ Herentals | 0 | 6 | 0 | 1 |
| AZ Turnhout | 32 | 0 | 0 | 0 |
| Protein Name | Gene Name | Number of Unique Peptides (seq. z) | Median Fold Change | FDR-Corrected p Value |
|---|---|---|---|---|
| Healthy controls vs. first diagnosis NMIBC | ||||
| Hemoglobin subunit alpha | HBA1 | 6 | 3.31 | 1.01 × 10−6 |
| Hemoglobin subunit beta | HBB | 9 | 3.21 | 2.76 × 10−5 |
| Alpha-2-macroglobulin | A2M | 35 | 2.14 | 3.03 × 10−15 |
| Fibrinogen gamma chain | FGG | 18 | 1.64 | 4.38 × 10−4 |
| Fibrinogen beta chain | FGB | 24 | 1.63 | 3.48 × 10−7 |
| Apolipoprotein A2 | APOA2 | 10 | 1.40 | 2.19 × 10−4 |
| Complement C3 | C3 | 65 | 1.27 | 2.08 × 10−21 |
| Apolipoprotein A1 | APOA1 | 29 | 1.25 | 1.12 × 10−11 |
| T-complex protein 1 subunit eta | CCT7 | 1 | 1.11 | 2.33 × 10−3 |
| Peroxiredoxin-2 | PRDX2 | 13 | 0.98 | 2.09 × 10−4 |
| Histone H4 | HIST1H4A | 6 | 0.95 | 5.57 × 10−4 |
| Alpha-1-antitrypsin | SERPINA1 | 24 | 0.82 | 6.40 × 10−5 |
| Alpha-1-antichymotrypsin | SERPINA3/GIG25 | 15 | 0.63 | 1.32 × 10−4 |
| Apolipoprotein D | APOD | 14 | −0.62 | 1.74 × 10−3 |
| Tumor susceptibility gene 101 protein | TSG101 | 16 | −0.63 | 1.56 × 10−2 |
| Prominin-1 | PROM1 | 32 | −0.64 | 1.73 × 10−4 |
| Galectin-3-binding protein | LGALS3BP | 19 | −0.64 | 5.10 × 10−5 |
| Annexin A11 | ANXA11 | 35 | −0.65 | 1.45 × 10−7 |
| Neprilysin | MME | 28 | −0.67 | 1.32 × 10−4 |
| Vacuolar-protein-sorting-associated protein 4A | VSP4A | 13 | −0.67 | 6.04 × 10−4 |
| Phospholipid scramblase 1 | PLSCR1 | 7 | −0.73 | 3.94 × 10−2 |
| Vacuolar-protein-sorting-associated protein 37D | VPS37D | 11 | −0.75 | 2.80 × 10−4 |
| Aminopeptidase N | ANPEP | 36 | −0.76 | 1.44 × 10−9 |
| Solute carrier family 12 member 1 | SLC12A1 | 26 | −0.76 | 1.32 × 10−4 |
| Sodium channel protein type 4 subunit alpha | SCN4A | 1 | −0.76 | 3.96 × 10−2 |
| E3 ubiquitin-protein ligase | MIB2 | 1 | −0.78 | 1.64 × 10−2 |
| MARCKS-related protein | MARCKSL1 | 1 | −0.79 | 2.19 × 10−2 |
| Cluster of differentiation 59 | CD59 | 14 | −0.80 | 1.83 × 10−6 |
| Aminoacylase-1 | ACY1 | 6 | −0.80 | 3.92 × 10−2 |
| Plasma serine protease inhibitor | SERPINA5 | 10 | −0.81 | 2.90 × 10−3 |
| Vacuolar-protein-sorting-associated protein 28 | VPS28 | 4 | −0.82 | 3.06 × 10−2 |
| Stomatin-like protein 3 | STOML3 | 1 | −0.82 | 1.26 × 10−2 |
| Coiled-coil domain-containing protein 168 | CCDC168 | 1 | −0.83 | 4.38 × 10−3 |
| Dipeptidase 1 | DPEP1 | 12 | −0.83 | 3.85 × 10−2 |
| Prostatic acid phosphatase | ACPP | 12 | −0.83 | 2.60 × 10−3 |
| Basal cell adhesion molecule | BCAM | 11 | −0.84 | 1.27 × 10−2 |
| Na(+)/H(+) exchange regulatory cofactor NHE-RF1 | NHE-RF1/SLC9A3R1 | 11 | −0.85 | 3.98 × 10−3 |
| Mucin-1 | MUC1 | 11 | −0.87 | 4.44 × 10−3 |
| Lipopolysaccharide-induced tumor necrosis factor-alpha factor | LITAF | 1 | −0.90 | 5.03 × 10−3 |
| Dipeptidyl peptidase 4 | DPP4 | 14 | −0.91 | 3.54 × 10−3 |
| Ubiquitin-fold modifier-conjugating enzyme 1 | UFC1 | 1 | −0.91 | 2.05 × 10−3 |
| BRO1 domain-containing protein | BROX | 15 | −0.96 | 1.01 × 10−4 |
| Protein tyrosine phosphatase receptor type J | PTPRJ | 16 | −0.96 | 6.71 × 10−5 |
| Charged multivesicular body protein 4b | CHMP4B | 9 | −0.97 | 7.67 × 10−4 |
| Kininogen-1 | KNG1 | 17 | −0.97 | 4.92 × 10−6 |
| Vasorin | VASN | 20 | −0.97 | 6.57 × 10−5 |
| IST1 homolog | IST1 | 18 | −0.98 | 2.67 × 10−4 |
| Pro-epidermal growth factor | EGF | 24 | −1.00 | 2.22 × 10−7 |
| Leucine-rich repeat-containing protein 71 | LRRC71 | 1 | −1.03 | 2.78 × 10−3 |
| Phosphatidylethanolamine-binding protein 1 | PEBP1 | 17 | −1.06 | 1.40 × 10−4 |
| Charged multivesicular body protein 2b | CHMP2B | 6 | −1.08 | 3.01 × 10−3 |
| Vesicular integral membrane protein VIP36 | LMAN2 | 13 | −1.09 | 2.22 × 10−4 |
| Uncharacterized protein C11orf52 | C11orf52 | 7 | −1.09 | 3.42 × 10−2 |
| G-protein coupled receptor 98 | GPR98 | 1 | −1.11 | 5.27 × 10−3 |
| Lysophosphatidylcholine acyltransferase 2 | LPCAT2 | 1 | −1.11 | 4.66 × 10−3 |
| Charged multivesicular body protein 2a | CHMP2A | 7 | −1.12 | 7.27 × 10−3 |
| Ubiquitin carboxyl-terminal hydrolase | UCHL1 | 1 | −1.12 | 1.34 × 10−3 |
| Mannosyl-oligosaccharide 1.2-alpha-mannosidase IA | MAN1A1 | 6 | −1.16 | 3.68 × 10−2 |
| E3 ubiquitin-protein ligase NEDD4 like | NEDD4L | 1 | −1.18 | 2.95 × 10−3 |
| HLA class II histocompatibility antigen. DM alpha chain | HLA-DMA | 1 | −1.21 | 2.78 × 10−3 |
| Collagen alpha-1(VI) chain | COL6A1 | 18 | −1.27 | 2.34 × 10−5 |
| Mannan-binding lectin serine protease 2 | MASP2 | 10 | −1.29 | 3.59 × 10−3 |
| Cytochrome b reductase 1 | CYBRD1 | 1 | −1.30 | 1.17 × 10−3 |
| Semaphorin-3G | SEMA3G | 1 | −1.32 | 2.49 × 10−3 |
| Small integral membrane protein 5 | SMIM5 | 1 | −1.33 | 1.26 × 10−3 |
| Fructose-bisphosphate aldolase B | ALDOB | 9 | −1.42 | 4.89 × 10−3 |
| Uromodulin | UMOD | 40 | −1.43 | 1.21 × 10−12 |
| Matrix-remodeling-associated protein 8 | MXRA8 | 12 | −1.45 | 4.58 × 10−5 |
| Gamma-glytamyl transpeptidase 1 | GGT1 | 1 | −1.80 | 2.15 × 10−4 |
| Tumor-free vs. recurrence NMBIC | ||||
| Hemoglobin subunit alpha | HBA1 | 6 | 1.10 | 3.19 × 10−3 |
| Hemoglobin subunit beta | HBB | 9 | 0.86 | 3.39 × 10−2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jordaens, S.; Oeyen, E.; Willems, H.; Ameye, F.; De Wachter, S.; Pauwels, P.; Mertens, I. Protein Biomarker Discovery Studies on Urinary sEV Fractions Separated with UF-SEC for the First Diagnosis and Detection of Recurrence in Bladder Cancer Patients. Biomolecules 2023, 13, 932. https://doi.org/10.3390/biom13060932
Jordaens S, Oeyen E, Willems H, Ameye F, De Wachter S, Pauwels P, Mertens I. Protein Biomarker Discovery Studies on Urinary sEV Fractions Separated with UF-SEC for the First Diagnosis and Detection of Recurrence in Bladder Cancer Patients. Biomolecules. 2023; 13(6):932. https://doi.org/10.3390/biom13060932
Chicago/Turabian StyleJordaens, Stephanie, Eline Oeyen, Hanny Willems, Filip Ameye, Stefan De Wachter, Patrick Pauwels, and Inge Mertens. 2023. "Protein Biomarker Discovery Studies on Urinary sEV Fractions Separated with UF-SEC for the First Diagnosis and Detection of Recurrence in Bladder Cancer Patients" Biomolecules 13, no. 6: 932. https://doi.org/10.3390/biom13060932
APA StyleJordaens, S., Oeyen, E., Willems, H., Ameye, F., De Wachter, S., Pauwels, P., & Mertens, I. (2023). Protein Biomarker Discovery Studies on Urinary sEV Fractions Separated with UF-SEC for the First Diagnosis and Detection of Recurrence in Bladder Cancer Patients. Biomolecules, 13(6), 932. https://doi.org/10.3390/biom13060932