Recent Advances in Peptidoglycan Synthesis and Regulation in Bacteria
Abstract
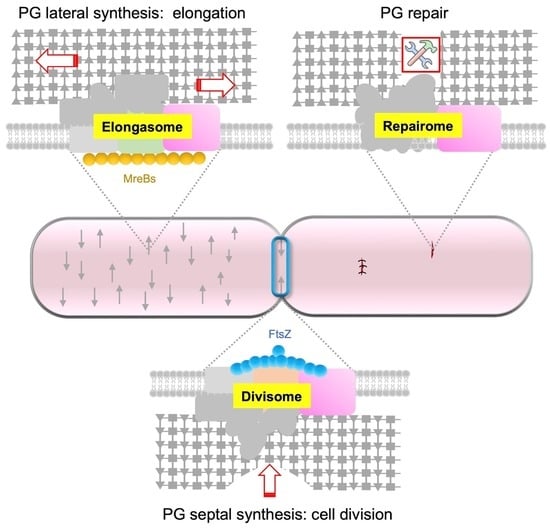
:1. Introduction
2. Regulation of GlcN-6-P Synthesis, the Initial Cytoplasmic Precursor of UDP-GlcNAc
2.1. Regulation of GlmS Synthesis in E. coli
2.2. Regulation of Synthesis and Activity of GlmS in B. subtilis
3. Flipping over the Cytoplasmic Membrane
3.1. The Lipid-II Flippases
3.2. The UndP Flippases
4. Regulation of PG Expansion
4.1. Regulation of PBPs
4.2. Regulation of PG Hydrolases
5. Respective Role of PBPs and SEDS
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Egan, A.J.F.; Errington, J.; Vollmer, W. Regulation of peptidoglycan synthesis and remodelling. Nat. Rev. Microbiol. 2020, 18, 446–460. [Google Scholar] [CrossRef] [PubMed]
- Errington, J.; Wu, L.J. Cell Cycle Machinery in Bacillus subtilis. Subcell. Biochem. 2017, 84, 67–101. [Google Scholar] [PubMed]
- Sassine, J.; Sousa, J.; Lalk, M.; Daniel, R.A.; Vollmer, W. Cell morphology maintenance in Bacillus subtilis through balanced peptidoglycan synthesis and hydrolysis. Sci. Rep. 2020, 10, 17910. [Google Scholar] [CrossRef] [PubMed]
- Weaver, A.; Taguchi, A.; Dörr, T. Masters of Misdirection: Peptidoglycan Glycosidases in Bacterial Growth. J. Bacteriol. 2023, 205, e0042822. [Google Scholar] [CrossRef]
- Brogan, A.P.; Rudner, D.Z. Regulation of peptidoglycan hydrolases: Localization, abundance, and activity. Curr. Opin. Microbiol. 2023, 72, 102279. [Google Scholar] [CrossRef]
- Kumar, S.; Mollo, A.; Kahne, D.; Ruiz, N. The Bacterial Cell Wall: From Lipid II Flipping to Polymerization. Chem. Rev. 2022, 122, 8884–8910. [Google Scholar] [CrossRef]
- Barreteau, H.; Kovac, A.; Boniface, A.; Sova, M.; Gobec, S.; Blanot, D. Cytoplasmic steps of peptidoglycan biosynthesis. FEMS Microbiol. Rev. 2008, 32, 168–207. [Google Scholar] [CrossRef]
- Mengin-Lecreulx, D.; Flouret, B.; van Heijenoort, J. Pool levels of UDP N-acetylglucosamine and UDP N-acetylglucosamine-enolpyruvate in Escherichia coli and correlation with peptidoglycan synthesis. J. Bacteriol. 1983, 154, 1284–1290. [Google Scholar] [CrossRef]
- Mengin-Lecreulx, D.; Flouret, B.; van Heijenoort, J. Cytoplasmic steps of peptidoglycan synthesis in Escherichia coli. J. Bacteriol. 1982, 151, 1109–1117. [Google Scholar] [CrossRef]
- Benson, T.E.; Walsh, C.T.; Hogle, J. The structure of the substrate-free form of MurB, an essential enzyme for the synthesis of bacterial cell walls. Structure 1996, 4, 47–54. [Google Scholar] [CrossRef]
- Brown, E.D.; Vivas, E.I.; Walsh, C.T.; Kolter, R. MurA (MurZ), the enzyme that catalyzes the first committed step in peptidoglycan biosynthesis, is essential in Escherichia coli. J. Bacteriol. 1995, 177, 4194–4197. [Google Scholar] [CrossRef] [PubMed]
- Vollmer, W.; Bertsche, U. Murein (peptidoglycan) structure, architecture and biosynthesis in Escherichia coli. Biochim. Biophys. Acta (BBA)-Biomembr. 2008, 1778, 1714–1734. [Google Scholar] [CrossRef]
- Morales Angeles, D.; Liu, Y.; Hartman, A.M.; Borisova, M.; de Sousa Borges, A.; de Kok, N.; Beilharz, K.; Veening, J.W.; Mayer, C.; Hirsch, A.K.; et al. Pentapeptide-rich peptidoglycan at the Bacillus subtilis cell-division site. Mol. Microbiol. 2017, 104, 319–333. [Google Scholar] [CrossRef] [PubMed]
- Bouhss, A.; Mengin-Lecreulx, D.; Le Beller, D.; Van Heijenoort, J. Topological analysis of the MraY protein catalysing the first membrane step of peptidoglycan synthesis. Mol. Microbiol. 1999, 34, 576–585. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, M.; Wachi, M.; Jung, H.K.; Ishino, F.; Matsuhashi, M. The Escherichia coli mraY gene encoding UDP-N-acetylmuramoyl-pentapeptide: Undecaprenyl-phosphate phospho-N-acetylmuramoyl-pentapeptide transferase. J. Bacteriol. 1991, 173, 1021–1026. [Google Scholar] [CrossRef]
- Lloyd, A.J.; Brandish, P.E.; Gilbey, A.M.; Bugg, T.D. Phospho-N-acetyl-muramyl-pentapeptide translocase from Escherichia coli: Catalytic role of conserved aspartic acid residues. J. Bacteriol. 2004, 186, 1747–1757. [Google Scholar] [CrossRef] [PubMed]
- Mengin-Lecreulx, D.; Texier, L.; Rousseau, M.; van Heijenoort, J. The murG gene of Escherichia coli codes for the UDP-N-acetylglucosamine: N-acetylmuramyl-(pentapeptide) pyrophosphoryl-undecaprenol N-acetylglucosamine transferase involved in the membrane steps of peptidoglycan synthesis. J. Bacteriol. 1991, 173, 4625–4636. [Google Scholar] [CrossRef]
- Ruiz, N. Bioinformatics identification of MurJ (MviN) as the peptidoglycan lipid II flippase in Escherichia coli. Proc. Natl. Acad. Sci. USA 2008, 105, 15553–15557. [Google Scholar] [CrossRef]
- Roney, I.J.; Rudner, D.Z. Two broadly conserved families of polyprenyl-phosphate transporters. Nature 2023, 613, 729–734. [Google Scholar] [CrossRef]
- Sit, B.; Srisuknimit, V.; Bueno, E.; Zingl, F.G.; Hullahalli, K.; Cava, F.; Waldor, M.K. Undecaprenyl phosphate translocases confer conditional microbial fitness. Nature 2023, 613, 721–728. [Google Scholar] [CrossRef]
- Szwedziak, P.; Löwe, J. Do the divisome and elongasome share a common evolutionary past? Curr. Opin. Microbiol. 2013, 16, 745–751. [Google Scholar] [CrossRef] [PubMed]
- Sharifzadeh, S.; Dempwolff, F.; Kearns, D.B.; Carlson, E.E. Harnessing β-Lactam Antibiotics for Illumination of the Activity of Penicillin-Binding Proteins in Bacillus subtilis. ACS Chem. Biol. 2020, 15, 1242–1251. [Google Scholar] [CrossRef] [PubMed]
- Hugonnet, J.E.; Mengin-Lecreulx, D.; Monton, A.; den Blaauwen, T.; Carbonnelle, E.; Veckerlé, C.; Brun, Y.V.; van Nieuwenhze, M.; Bouchier, C.; Tu, K.; et al. Factors essential for L,D-transpeptidase-mediated peptidoglycan cross-linking and β-lactam resistance in Escherichia coli. Elife 2016, 5, e19469. [Google Scholar] [CrossRef] [PubMed]
- Goffin, C.; Ghuysen, J.M. Multimodular penicillin-binding proteins: An enigmatic family of orthologs and paralogs. Microbiol. Mol. Biol. Rev. 1998, 62, 1079–1093. [Google Scholar] [CrossRef]
- Sjodt, M.; Rohs, P.D.A.; Gilman, M.S.A.; Erlandson, S.C.; Zheng, S.; Green, A.G.; Brock, K.P.; Taguchi, A.; Kahne, D.; Walker, S.; et al. Structural coordination of polymerization and crosslinking by a SEDS-bPBP peptidoglycan synthase complex. Nat. Microbiol. 2020, 5, 813–820. [Google Scholar] [CrossRef]
- Emami, K.; Guyet, A.; Kawai, Y.; Devi, J.; Wu, L.J.; Allenby, N.; Daniel, R.A.; Errington, J. RodA as the missing glycosyltransferase in Bacillus subtilis and antibiotic discovery for the peptidoglycan polymerase pathway. Nat. Microbiol. 2017, 2, 16253. [Google Scholar] [CrossRef]
- Meeske, A.J.; Riley, E.P.; Robins, W.P.; Uehara, T.; Mekalanos, J.J.; Kahne, D.; Walker, S.; Kruse, A.C.; Bernhardt, T.G.; Rudner, D.Z. SEDS proteins are a widespread family of bacterial cell wall polymerases. Nature 2016, 537, 634–638. [Google Scholar] [CrossRef] [PubMed]
- Carballido-López, R.; Formstone, A.; Li, Y.; Ehrlich, S.D.; Noirot, P.; Errington, J. Actin homolog MreBH governs cell morphogenesis by localization of the cell wall hydrolase LytE. Dev. Cell 2006, 11, 399–409. [Google Scholar] [CrossRef]
- Bohrhunter, J.L.; Rohs, P.D.A.; Torres, G.; Yunck, R.; Bernhardt, T.G. MltG activity antagonizes cell wall synthesis by both types of peptidoglycan polymerases in Escherichia coli. Mol. Microbiol. 2021, 115, 1170–1180. [Google Scholar] [CrossRef]
- Sassine, J.; Pazos, M.; Breukink, E.; Vollmer, W. Lytic transglycosylase MltG cleaves in nascent peptidoglycan and produces short glycan strands. Cell Surf. 2021, 7, 100053. [Google Scholar] [CrossRef]
- Mengin-Lecreulx, D.; van Heijenoort, J. Identification of the glmU gene encoding N-acetylglucosamine-1-phosphate uridyltransferase in Escherichia coli. J. Bacteriol. 1993, 175, 6150–6157. [Google Scholar] [CrossRef]
- Winkler, W.C.; Nahvi, A.; Roth, A.; Collins, J.A.; Breaker, R.R. Control of gene expression by a natural metabolite-responsive ribozyme. Nature 2004, 428, 281–286. [Google Scholar] [CrossRef] [PubMed]
- Kalamorz, F.; Reichenbach, B.; März, W.; Rak, B.; Görke, B. Feedback control of glucosamine-6-phosphate synthase GlmS expression depends on the small RNA GlmZ and involves the novel protein YhbJ in Escherichia coli. Mol. Microbiol. 2007, 65, 1518–1533. [Google Scholar] [CrossRef] [PubMed]
- Foulquier, E.; Pompeo, F.; Byrne, D.; Fierobe, H.P.; Galinier, A. Uridine diphosphate N-acetylglucosamine orchestrates the interaction of GlmR with either YvcJ or GlmS in Bacillus subtilis. Sci. Rep. 2020, 10, 15938. [Google Scholar] [CrossRef] [PubMed]
- Luciano, J.; Foulquier, E.; Fantino, J.R.; Galinier, A.; Pompeo, F. Characterization of YvcJ, a conserved P-loop-containing protein, and its implication in competence in Bacillus subtilis. J. Bacteriol. 2009, 191, 1556–1564. [Google Scholar] [CrossRef] [PubMed]
- Durica-Mitic, S.; Görke, B. Feedback regulation of small RNA processing by the cleavage product. RNA Biol. 2019, 16, 1055–1065. [Google Scholar] [CrossRef] [PubMed]
- Göpel, Y.; Görke, B. Interaction of lipoprotein QseG with sensor kinase QseE in the periplasm controls the phosphorylation state of the two-component system QseE/QseF in Escherichia coli. PLoS Genet. 2018, 14, e1007547. [Google Scholar] [CrossRef]
- Göpel, Y.; Khan, M.A.; Görke, B. Ménage à trois: Post-transcriptional control of the key enzyme for cell envelope synthesis by a base-pairing small RNA, an RNase adaptor protein, and a small RNA mimic. RNA Biol. 2014, 11, 433–442. [Google Scholar] [CrossRef]
- Khan, M.A.; Durica-Mitic, S.; Göpel, Y.; Heermann, R.; Görke, B. Small RNA-binding protein RapZ mediates cell envelope precursor sensing and signaling in Escherichia coli. EMBO J. 2020, 39, e103848. [Google Scholar] [CrossRef]
- Islam, M.S.; Hardwick, S.W.; Quell, L.; Durica-Mitic, S.; Chirgadze, D.Y.; Görke, B.; Luisi, B.F. Structure of a bacterial ribonucleoprotein complex central to the control of cell envelope biogenesis. EMBO J. 2022, e112574. [Google Scholar] [CrossRef]
- McCown, P.J.; Winkler, W.C.; Breaker, R.R. Mechanism and distribution of glmS ribozymes. Methods Mol. Biol. 2012, 848, 113–129. [Google Scholar] [PubMed]
- Winkler, W.C.; Breaker, R.R. Regulation of bacterial gene expression by riboswitches. Annu. Rev. Microbiol. 2005, 59, 487–517. [Google Scholar] [CrossRef] [PubMed]
- Cochrane, J.C.; Lipchock, S.V.; Smith, K.D.; Strobel, S.A. Structural and chemical basis for glucosamine 6-phosphate binding and activation of the glmS ribozyme. Biochemistry 2009, 48, 3239–3246. [Google Scholar] [CrossRef] [PubMed]
- Cochrane, J.C.; Strobel, S.A. Catalytic strategies of self-cleaving ribozymes. Acc. Chem. Res. 2008, 41, 1027–1035. [Google Scholar] [CrossRef]
- Patel, V.; Wu, Q.; Chandrangsu, P.; Helmann, J.D. A metabolic checkpoint protein GlmR is important for diverting carbon into peptidoglycan biosynthesis in Bacillus subtilis. PLoS Genet. 2018, 14, e1007689. [Google Scholar] [CrossRef]
- Görke, B.; Foulquier, E.; Galinier, A. YvcK of Bacillus subtilis is required for a normal cell shape and for growth on Krebs cycle intermediates and substrates of the pentose phosphate pathway. Microbiology 2005, 151 Pt 11, 3777–3791. [Google Scholar] [CrossRef]
- Foulquier, E.; Galinier, A. YvcK, a protein required for cell wall integrity and optimal carbon source utilization, binds uridine diphosphate-sugars. Sci. Rep. 2017, 7, 4139. [Google Scholar] [CrossRef]
- Pompeo, F.; Luciano, J.; Brochier-Armanet, C.; Galinier, A. The GTPase function of YvcJ and its subcellular relocalization are dependent on growth conditions in Bacillus subtilis. J. Mol. Microbiol. Biotechnol. 2011, 20, 156–167. [Google Scholar] [CrossRef]
- Patel, Y.; Soni, V.; Rhee, K.Y.; Helmann, J.D. Mutations in rpoB That Confer Rifampicin Resistance Can Alter Levels of Peptidoglycan Precursors and Affect β-Lactam Susceptibility. mBio 2023, e0316822. [Google Scholar] [CrossRef]
- Kumar, S.; Rubino, F.A.; Mendoza, A.G.; Ruiz, N. The bacterial lipid II flippase MurJ functions by an alternating-access mechanism. J. Biol. Chem. 2019, 294, 981–990. [Google Scholar] [CrossRef]
- Rubino, F.A.; Kumar, S.; Ruiz, N.; Walker, S.; Kahne, D.E. Membrane Potential Is Required for MurJ Function. J. Am. Chem. Soc. 2018, 140, 4481–4484. [Google Scholar] [CrossRef] [PubMed]
- Fay, A.; Dworkin, J. Bacillus subtilis homologs of MviN (MurJ), the putative Escherichia coli lipid II flippase, are not essential for growth. J. Bacteriol. 2009, 191, 6020–6028. [Google Scholar] [CrossRef] [PubMed]
- Meeske, A.J.; Sham, L.T.; Kimsey, H.; Koo, B.M.; Gross, C.A.; Bernhardt, T.G.; Rudner, D.Z. MurJ and a novel lipid II flippase are required for cell wall biogenesis in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 2015, 112, 6437–6442. [Google Scholar] [CrossRef]
- Mohammadi, T.; van Dam, V.; Sijbrandi, R.; Vernet, T.; Zapun, A.; Bouhss, A.; Diepeveen-de Bruin, M.; Nguyen-Distèche, M.; de Kruijff, B.; Breukink, E. Identification of FtsW as a transporter of lipid-linked cell wall precursors across the membrane. EMBO J. 2011, 30, 1425–1432. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Kumar, S.; Ruiz, N.; Walker, S.; Kahne, D.E. FtsW activity and lipid II synthesis are required for recruitment of MurJ to midcell during cell division in Escherichia coli. Mol. Microbiol. 2018, 109, 855–884. [Google Scholar] [CrossRef] [PubMed]
- Manat, G.; El Ghachi, M.; Auger, R.; Baouche, K.; Olatunji, S.; Kerff, F.; Touzé, T.; Mengin-Lecreulx, D.; Bouhss, A. Membrane Topology and Biochemical Characterization of the Escherichia coli BacA Undecaprenyl-Pyrophosphate Phosphatase. PLoS ONE 2015, 10, e0142870. [Google Scholar] [CrossRef]
- Zhao, H.; Sun, Y.; Peters, J.M.; Gross, C.A.; Garner, E.C.; Helmann, J.D. Depletion of Undecaprenyl Pyrophosphate Phosphatases Disrupts Cell Envelope Biogenesis in Bacillus subtilis. J. Bacteriol. 2016, 198, 2925–2935. [Google Scholar] [CrossRef]
- Garner, E.C.; Bernard, R.; Wang, W.; Zhuang, X.; Rudner, D.Z.; Mitchison, T. Coupled, circumferential motions of the cell wall synthesis machinery and MreB filaments in B. subtilis. Science 2011, 333, 222–225. [Google Scholar] [CrossRef]
- Domínguez-Escobar, J.; Chastanet, A.; Crevenna, A.H.; Fromion, V.; Wedlich-Söldner, R.; Carballido-López, R. Processive movement of MreB-associated cell wall biosynthetic complexes in bacteria. Science 2011, 333, 225–228. [Google Scholar] [CrossRef]
- Patel, Y.; Zhao, H.; Helmann, J.D. A regulatory pathway that selectively up-regulates elongasome function in the absence of class A PBPs. Elife 2020, 9, e57902. [Google Scholar] [CrossRef]
- Errington, J. Bacterial morphogenesis and the enigmatic MreB helix. Nat. Rev. Microbiol. 2015, 13, 241–248. [Google Scholar] [CrossRef] [PubMed]
- de Boer, P.; Crossley, R.; Rothfield, L. The essential bacterial cell-division protein FtsZ is a GTPase. Nature 1992, 359, 254–256. [Google Scholar] [CrossRef] [PubMed]
- RayChaudhuri, D.; Park, J.T. Escherichia coli cell-division gene ftsZ encodes a novel GTP-binding protein. Nature 1992, 359, 251–254. [Google Scholar] [CrossRef] [PubMed]
- Gamba, P.; Veening, J.W.; Saunders, N.J.; Hamoen, L.W.; Daniel, R.A. Two-step assembly dynamics of the Bacillus subtilis divisome. J. Bacteriol. 2009, 191, 4186–4194. [Google Scholar] [CrossRef] [PubMed]
- Du, S.; Lutkenhaus, J. Assembly and activation of the Escherichia coli divisome. Mol. Microbiol. 2017, 105, 177–187. [Google Scholar] [CrossRef] [PubMed]
- Navarro, P.P.; Vettiger, A.; Ananda, V.Y.; Llopis, P.M.; Allolio, C.; Bernhardt, T.G.; Chao, L.H. Cell wall synthesis and remodelling dynamics determine division site architecture and cell shape in Escherichia coli. Nat. Microbiol. 2022, 7, 1621–1634. [Google Scholar] [CrossRef] [PubMed]
- den Blaauwen, T.; de Pedro, M.A.; Nguyen-Distèche, M.; Ayala, J.A. Morphogenesis of rod-shaped sacculi. FEMS Microbiol. Rev. 2008, 32, 321–344. [Google Scholar] [CrossRef]
- Sassine, J.; Xu, M.; Sidiq, K.R.; Emmins, R.; Errington, J.; Daniel, R.A. Functional redundancy of division specific penicillin-binding proteins in Bacillus subtilis. Mol. Microbiol. 2017, 106, 304–318. [Google Scholar] [CrossRef]
- Wei, Y.; Havasy, T.; McPherson, D.C.; Popham, D.L. Rod shape determination by the Bacillus subtilis class B penicillin-binding proteins encoded by pbpA and pbpH. J. Bacteriol. 2003, 185, 4717–4726. [Google Scholar] [CrossRef]
- Daniel, R.A.; Harry, E.J.; Errington, J. Role of penicillin-binding protein PBP 2B in assembly and functioning of the division machinery of Bacillus subtilis. Mol. Microbiol. 2000, 35, 299–311. [Google Scholar] [CrossRef]
- McPherson, D.C.; Driks, A.; Popham, D.L. Two class A high-molecular-weight penicillin-binding proteins of Bacillus subtilis play redundant roles in sporulation. J. Bacteriol. 2001, 183, 6046–6053. [Google Scholar] [CrossRef] [PubMed]
- Claessen, D.; Emmins, R.; Hamoen, L.W.; Daniel, R.A.; Errington, J.; Edwards, D.H. Control of the cell elongation-division cycle by shuttling of PBP1 protein in Bacillus subtilis. Mol. Microbiol. 2008, 68, 1029–1046. [Google Scholar] [CrossRef] [PubMed]
- Cleverley, R.M.; Rutter, Z.J.; Rismondo, J.; Corona, F.; Tsui, H.T.; Alatawi, F.A.; Daniel, R.A.; Halbedel, S.; Massidda, O.; Winkler, M.E.; et al. The cell cycle regulator GpsB functios as cytosolic adaptor for multiple cell wall enzymes. Nat. Commun. 2019, 10, 261. [Google Scholar] [CrossRef] [PubMed]
- Paradis-Bleau, C.; Markovski, M.; Uehara, T.; Lupoli, T.J.; Walker, S.; Kahne, D.E.; Bernhardt, T.G. Lipoprotein cofactors located in the outer membrane activate bacterial cell wall polymerases. Cell 2010, 143, 1110–1120. [Google Scholar] [CrossRef]
- Typas, A.; Banzhaf, M.; van den Berg van Saparoea, B.; Verheul, J.; Biboy, J.; Nichols, R.J.; Zietek, M.; Beilharz, K.; Kannenberg, K.; von Rechenberg, M.; et al. Regulation of peptidoglycan synthesis by outer-membrane proteins. Cell 2010, 143, 1097–1109. [Google Scholar] [CrossRef]
- Sardis, M.F.; Bohrhunter, J.L.; Greene, N.G.; Bernhardt, T.G. The LpoA activator is required to stimulate the peptidoglycan polymerase activity of its cognate cell wall synthase PBP1a. Proc. Natl. Acad. Sci. USA 2021, 118, e210889411. [Google Scholar] [CrossRef] [PubMed]
- Kermani, A.A.; Biboy, J.; Vollmer, D.; Vollmer, W. Outer membrane-anchoring enables LpoB to regulate peptidoglycan synthesis rate. Cell Surf. 2022, 8, 100086. [Google Scholar] [CrossRef]
- Delisle, J.; Cordier, B.; Audebert, S.; Pophillat, M.; Cluzel, C.; Espinosa, L.; Grangeasse, C.; Galinier, A.; Doan, T. Characterization of TseB: A new actor in cell wall elongation in Bacillus subtilis. Mol. Microbiol. 2021, 116, 1099–1112. [Google Scholar] [CrossRef]
- Stamsås, G.A.; Myrbråten, I.S.; Straume, D.; Salehian, Z.; Veening, J.W.; Håvarstein, L.S.; Kjos, M. CozEa and CozEb play overlapping and essential roles in controlling cell division in Staphylococcus aureus. Mol. Microbiol. 2018, 109, 615–632. [Google Scholar] [CrossRef]
- Fenton, A.K.; El Mortaji, L.; Lau, D.T.; Rudner, D.Z.; Bernhardt, T.G. CozE is a member of the MreCD complex that directs cell elongation in Streptococcus pneumoniae. Nat. Microbiol. 2016, 2, 16237. [Google Scholar] [CrossRef]
- Fenton, A.K.; Manuse, S.; Flores-Kim, J.; Garcia, P.S.; Mercy, C.; Grangeasse, C.; Bernhardt, T.G.; Rudner, D.Z. Phosphorylation-dependent activation of the cell wall synthase PBP2a in Streptococcus pneumoniae by MacP. Proc. Natl. Acad. Sci. USA 2018, 115, 2812–2817. [Google Scholar] [CrossRef] [PubMed]
- Winther, A.R.; Kjos, M.; Herigstad, M.L.; Håvarstein, L.S.; Straume, D. EloR interacts with the lytic transglycosylase MltG at midcell in Streptococcus pneumoniae R6. J. Bacteriol. 2021, 203, e00691-20. [Google Scholar] [CrossRef] [PubMed]
- Stamsås, G.A.; Straume, D.; Ruud Winther, A.; Kjos, M.; Frantzen, C.A.; Håvarstein, L.S. Identification of EloR (Spr1851) as a regulator of cell elongation in Streptococcus pneumoniae. Mol. Microbiol. 2017, 105, 954–967. [Google Scholar] [CrossRef]
- Hashimoto, M.; Ooiwa, S.; Sekiguchi, J. Synthetic lethality of the lytE cwlO genotype in Bacillus subtilis is caused by lack of D,L-endopeptidase activity at the lateral cell wall. J. Bacteriol. 2012, 194, 796–803. [Google Scholar] [CrossRef]
- Dobihal, G.S.; Flores-Kim, J.; Roney, I.J.; Wang, X.; Rudner, D.Z. The WalR-WalK Signaling Pathway Modulates the Activities of both CwlO and LytE through Control of the Peptidoglycan Deacetylase PdaC in Bacillus subtilis. J. Bacteriol. 2022, 204, e0053321. [Google Scholar] [CrossRef]
- Brunet, Y.R.; Wang, X.; Rudner, D.Z. SweC and SweD are essential co-factors of the FtsEX-CwlO cell wall hydrolase complex in Bacillus subtilis. PLoS Genet. 2019, 15, e1008296. [Google Scholar] [CrossRef]
- Du, S.; Pichoff, S.; Lutkenhaus, J. FtsEX acts on FtsA to regulate divisome assembly and activity. Proc. Natl. Acad. Sci. USA 2016, 113, E5052–E5061. [Google Scholar] [CrossRef]
- Du, S.; Henke, W.; Pichoff, S.; Lutkenhaus, J. How FtsEX localizes to the Z ring and interacts with FtsA to regulate cell division. Mol. Microbiol. 2019, 112, 881–895. [Google Scholar] [CrossRef]
- Straume, D.; Piechowiak, K.W.; Kjos, M.; Håvarstein, L.S. Class A PBPs: It is time to rethink traditional paradigms. Mol. Microbiol. 2021, 116, 41–52. [Google Scholar] [CrossRef]
- McPherson, D.C.; Popham, D.L. Peptidoglycan synthesis in the absence of class A penicillin-binding proteins in Bacillus subtilis. J. Bacteriol. 2003, 185, 1423–1431. [Google Scholar] [CrossRef] [PubMed]
- Pazos, M.; Vollmer, W. Regulation and function of class A Penicillin-binding proteins. Curr. Opin. Microbiol. 2021, 60, 80–87. [Google Scholar] [CrossRef] [PubMed]
- Welsh, M.A.; Schaefer, K.; Taguchi, A.; Kahne, D.; Walker, S. Direction of Chain Growth and Substrate Preferences of Shape, Elongation, Division, and Sporulation-Family Peptidoglycan Glycosyltransferases. J. Am. Chem. Soc. 2019, 141, 12994–12997. [Google Scholar] [CrossRef]
- Taguchi, A.; Welsh, M.A.; Marmont, L.S.; Lee, W.; Sjodt, M.; Kruse, A.C.; Kahne, D.; Bernhardt, T.G.; Walker, S. FtsW is a peptidoglycan polymerase that is functional only in complex with its cognate penicillin-binding protein. Nat. Microbiol. 2019, 4, 587–594. [Google Scholar] [CrossRef] [PubMed]
- Boylan, R.J.; Mendelson, N.H. Initial characterization of a temperature-sensitive rod--mutant of Bacillus subtilis. J. Bacteriol. 1969, 100, 1316–1321. [Google Scholar] [CrossRef]
- Marmont, L.S.; Bernhardt, T.G. A conserved subcomplex within the bacterial cytokinetic ring activates cell wall synthesis by the FtsW-FtsI synthase. Proc. Natl. Acad. Sci. USA 2020, 117, 23879–23885. [Google Scholar] [CrossRef]
- Yang, X.; McQuillen, R.; Lyu, Z.; Phillips-Mason, P.; De La Cruz, A.; McCausland, J.W.; Liang, H.; DeMeester, K.E.; Santiago, C.C.; Grimes, C.L.; et al. A two-track model for the spatiotemporal coordination of bacterial septal cell wall synthesis revealed by single-molecule imaging of FtsW. Nat. Microbiol. 2021, 6, 584–593. [Google Scholar] [CrossRef] [PubMed]
- Vigouroux, A.; Cordier, B.; Aristov, A.; Alvarez, L.; Özbaykal, G.; Chaze, T.; Oldewurtel, E.R.; Matondo, M.; Cava, F.; Bikard, D.; et al. Class-A penicillin binding proteins do not contribute to cell shape but repair cell-wall defects. Elife 2020, 9, e51998. [Google Scholar] [CrossRef]
- Dion, M.F.; Kapoor, M.; Sun, Y.; Wilson, S.; Ryan, J.; Vigouroux, A.; van Teeffelen, S.; Oldenbourg, R.; Garner, E.C. Bacillus subtilis cell diameter is determined by the opposing actions of two distinct cell wall synthetic systems. Nat. Microbiol. 2019, 4, 1294–1305. [Google Scholar] [CrossRef]
- Brunet, Y.R.; Habib, C.; Brogan, A.P.; Artzi, L.; Rudner, D.Z. Intrinsically disordered protein regions are required for cell wall homeostasis in Bacillus subtilis. Genes Dev. 2022, 36, 970–984. [Google Scholar]
- Straume, D.; Piechowiak, K.W.; Olsen, S.; Stamsås, G.A.; Berg, K.H.; Kjos, M.; Heggenhougen, M.V.; Alcorlo, M.; Hermoso, J.A.; Håvarstein, L.S. Class A PBPs have a distinct and unique role in the construction of the pneumococcal cell wall. Proc. Natl. Acad. Sci. USA 2020, 117, 6129–6138. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Galinier, A.; Delan-Forino, C.; Foulquier, E.; Lakhal, H.; Pompeo, F. Recent Advances in Peptidoglycan Synthesis and Regulation in Bacteria. Biomolecules 2023, 13, 720. https://doi.org/10.3390/biom13050720
Galinier A, Delan-Forino C, Foulquier E, Lakhal H, Pompeo F. Recent Advances in Peptidoglycan Synthesis and Regulation in Bacteria. Biomolecules. 2023; 13(5):720. https://doi.org/10.3390/biom13050720
Chicago/Turabian StyleGalinier, Anne, Clémentine Delan-Forino, Elodie Foulquier, Hakima Lakhal, and Frédérique Pompeo. 2023. "Recent Advances in Peptidoglycan Synthesis and Regulation in Bacteria" Biomolecules 13, no. 5: 720. https://doi.org/10.3390/biom13050720
APA StyleGalinier, A., Delan-Forino, C., Foulquier, E., Lakhal, H., & Pompeo, F. (2023). Recent Advances in Peptidoglycan Synthesis and Regulation in Bacteria. Biomolecules, 13(5), 720. https://doi.org/10.3390/biom13050720