An Amyloidogenic Sequence at the N-Terminus of the Androgen Receptor Impacts Polyglutamine Aggregation
Abstract
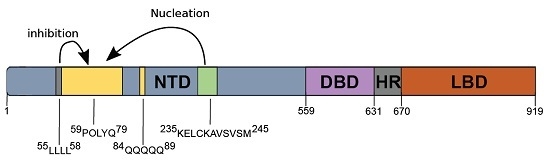
:1. Introduction
2. Results
2.1. Amyloidogenic Properties of KELCKAVSVSM Peptides Expressed as SUMO Fusion Proteins
2.2. The KELCKAVSVSM Sequence Modulates polyQ Oligomerization Properties of AR-NTD Fragments
3. Discussion
4. Materials and Methods
4.1. Cloning, Protein Expression, and Purification
4.2. Size Exclusion Chromatography Analysis of SUMO–Peptide Fusion
4.3. NMR Experiments and Formation of Amyloid Fibrils of KELCKAV Peptides
4.4. AFM Measurements of ARNTD Constructs
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Matias, P.M.; Donner, P.; Coelho, R.; Thomaz, M.; Peixoto, C.; Macedo, S.; Otto, N.; Joschko, S.; Scholz, P.; Wegg, A.; et al. Carrondo, Structural evidence for ligand specificity in the binding domain of the human androgen receptor: Implications for pathogenic gene mutations. J. Biol. Chem. 2000, 275, 26164–26171. [Google Scholar] [CrossRef] [PubMed]
- He, B.; Gampe, R.T., Jr.; Kole, A.J.; Hnat, A.T.; Stanley, T.B.; An, G.; Stewart, E.L.; Kalman, R.I.; Minges, J.T.; Wilson, E.M. Structural basis for androgen receptor interdomain and coactivator interactions suggests a transition in nuclear receptor activation function dominance. Mol. Cell 2004, 16, 425–438. [Google Scholar] [CrossRef] [PubMed]
- PShaffer, L.; Jivan, A.; Dollins, D.E.; Claessens, F.; Gewirth, D.T. Structural basis of androgen receptor binding to selective androgen response elements. Proc. Natl. Acad. Sci. USA 2004, 101, 4758–4763. [Google Scholar] [CrossRef] [PubMed]
- Lavery, D.N.; McEwan, I.J. Structural characterization of the native NH2-terminal transactivation domain of the human androgen receptor: A collapsed disordered conformation underlies structural plasticity and protein-induced folding. Biochemistry 2008, 47, 3360–3369. [Google Scholar] [CrossRef] [PubMed]
- Jenster, G.; van der Korput, H.A.; van Vroonhoven, C.; van der Kwast, T.H.; Trapman, J.; Brinkmann, A.O. Domains of the human androgen receptor involved in steroid binding, transcriptional activation, and subcellular localization. Mol. Endocrinol. 1991, 5, 1396–1404. [Google Scholar] [CrossRef] [PubMed]
- Simental, J.A.; Sar, M.; Lane, M.V.; French, F.V.; Wilson, E.M. Transcriptional activation and nuclear targeting signals of the human androgen receptor. J. Biol. Chem. 1991, 266, 510–518. [Google Scholar] [PubMed]
- Karlin, S.; Burge, C. Trinucleotide repeats and long homopeptides in genes and proteins associated with nervous system disease and development. Proc. Natl. Acad. Sci. USA 1996, 93, 1560–1565. [Google Scholar] [CrossRef] [PubMed]
- La Spada, A.R.; Wilson, E.M.; Lubahn, D.B.; Harding, A.E.; Fischbeck, K.H. Androgen receptor gene mutations in X-linked spinal and bulbar muscular atrophy. Nature 1991, 352, 77–79. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Atamna, H.; Zakharov, M.N.; Bhasin, S.; Khan, S.H.; Jasuja, R. Role of the androgen receptor CAG repeat polymorphism in prostate cancer, and spinal and bulbar muscular atrophy. Life Sci. 2011, 88, 565–571. [Google Scholar] [CrossRef] [PubMed]
- Jochum, T.; Ritz, M.E.; Schuster, C.; Funderburk, S.F.; Jehle, K.; Schmitz, K.; Brinkmann, F.; Hirtz, M.; Moss, D.; Cato, A.C.B. Toxic and non-toxic aggregates from the SBMA and normal forms of androgen receptor have distinct oligomeric structures. Biochim. Biophys. Acta (BBA) 2012, 1822, 1070–1078. [Google Scholar] [CrossRef] [PubMed]
- Duennwald, M.L.; Jagadish, S.; Muchowski, P.J.; Lindquist, S. Flanking sequences profoundly alter polyglutamine toxicity in yeast. Proc. Natl. Acad. Sci. USA 2006, 103, 11045–11050. [Google Scholar] [CrossRef] [PubMed]
- Eftekharzadeh, B.; Piai, A.; Chiesa, G.; Mungianu, D.; Garcia, J.; Pierattelli, R.; Felli, I.C.; Salvatella, X. Sequence Context Influences the Structure and Aggregation Behavior of a PolyQ Tract. Biophys. J. 2016, 110, 2361–2366. [Google Scholar] [CrossRef] [PubMed]
- Saunders, H.M.; Bottomley, S.P. Multi-domain misfolding: Understanding the aggregation pathway of polyglutamine proteins. Protein Eng. Des. Sel. 2009, 22, 447–451. [Google Scholar] [CrossRef] [PubMed]
- Masino, L.; Nicastro, G.; Menon, R.P.; Piaz, F.D.; Calder, L.; Pastore, A. Characterization of the structure and the amyloidogenic properties of the Josephin domain of the polyglutamine-containing protein ataxin-3. J. Mol. Biol. 2004, 344, 1021–1035. [Google Scholar] [CrossRef] [PubMed]
- De Chiara, C.; Menon, R.P.; Piaz, F.D.; Calder, L.; Pastore, A. Polyglutamine is not all: The functional role of the AXH domain in the ataxin-1 protein. J. Mol. Biol. 2005, 354, 883–893. [Google Scholar] [CrossRef] [PubMed]
- Asencio-Hernández, J.; Ruhlmann, C.; McEwen, A.; Eberling, P.; Nominé, Y.; Céraline, J.; Starck, J.-P.; Delsuc, M.A. Reversible amyloid fiber formation in the N terminus of androgen receptor. Chembiochem 2014, 15, 2370–2373. [Google Scholar] [CrossRef] [PubMed]
- Butt, T.R.; Edavettal, S.C.; Hall, J.P.; Mattern, M.R. SUMO fusion technology for difficult-to-express proteins. Protein Expr. Purif. 2005, 43, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Tam, J.P.; Wu, C.R.; Liu, W.; Zhang, J.W. Disulfide bond formation in peptides by dimethyl sulfoxide. Scope and applications. J. Am. Chem. Soc. 1991, 113, 6657–6662. [Google Scholar] [CrossRef]
- Kuiper, E.F.; de Mattos, E.P.; Jardim, L.B.; Kampinga, H.H.; Bergink, S. Chaperones in polyglutamine aggregation: Beyond the Q-stretch. Front. Neurosci. 2017, 11, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Margolis, R.L.; Ross, C.A. Expansion explosion: New clues to the pathogenesis of repeat expansion neurodegenerative diseases. Trends Mol. Med. 2001, 7, 479–482. [Google Scholar] [CrossRef]
- Crick, S.L.; Ruff, K.M.; Garai, K.; Frieden, C.; Pappu, R.V. Unmasking the roles of N- and C-terminal flanking sequences from exon 1 of huntingtin as modulators of polyglutamine aggregation. Proc. Natl. Acad. Sci. USA 2013, 110, 20075–20080. [Google Scholar] [CrossRef] [PubMed]
- Thakur, A.K.; Jayaraman, M.; Mishra, R.; Thakur, M.; Chellgren, V.M.; Byeon, I.-J.; Anjum, D.H.; Kodali, R.; Creamer, T.P.; Conway, J.F.; et al. Polyglutamine disruption of the huntingtin exon 1 N terminus triggers a complex aggregation mechanism. Nat. Struct. Mol. Biol. 2009, 16, 380–389. [Google Scholar] [CrossRef] [PubMed]
- De Mol, E.; Fenwick, R.B.; Phang, C.T.; Buzón, V.; Szulc, E.; de la Fuente, A.; Escobedo, A.; García, J.; Bertoncini, I.J.; Estébanez-Perpiñá, C.W.; et al. EPI-001, A Compound Active against Castration-Resistant Prostate Cancer, Targets Transactivation Unit 5 of the Androgen Receptor. ACS Chem. Biol. 2016. [Google Scholar] [CrossRef] [PubMed]
- Betney, R.; McEwan, I.J. Role of conserved hydrophobic amino acids in androgen receptor AF-1 function. J. Mol. Endocrinol. 2003, 31, 427–439. [Google Scholar] [CrossRef] [PubMed]
- He, B.; Bai, S.; Hnat, A.; Kalman, R.; Minges, J.; Patterson, C.; Wilson, E. An androgen receptor NH2-terminal conserved motif interacts with the COOH terminus of the Hsp70-interacting protein (CHIP). J. Biol. Chem. 2004, 279, 30643–30653. [Google Scholar] [CrossRef] [PubMed]
- Myung, J.K.; Wang, G.; Chiu, H.H.; Wang, J.; Mawji, N.R.; Sadar, M.D. Inhibition of androgen receptor by decoy molecules delays progression to castration-recurrent prostate cancer. PLoS ONE 2017, 12, e0174134. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Zapien, D.; Delsuc, M.A.; Travé, G.; Lutzing, R.; Rochette-Egly, C.; Kieffer, B. Production and characterization of a retinoic acid receptor RARγ construction encompassing the DNA binding domain and the disordered N-terminal proline rich domain. Protein Expr. Purif. 2014, 95, 113–120. [Google Scholar] [CrossRef] [PubMed]
- Meyer, S.; Wang, Y.H.; Pérez-Escrivà, P.; Kieffer, B. Backbone 1H, 15N, 13C NMR assignment of the 518–627 fragment of the androgen receptor encompassing N-terminal and DNA binding domains. Biomol. NMR Assign. 2016, 10, 175–178. [Google Scholar]
- Bax, A.; Grzesiek, S. Methodological advances in protein NMR. Acc. Chem. Res. 1993, 26, 131–138. [Google Scholar] [CrossRef]
- Wishart, D.S.; Bigam, C.G.; Yao, J.; Abildgaard, F.; Dyson, H.J.; Oldfield, E.; Markley, J.L.; Sykes, B.D. 1H, 13C and 15N chemical shift referencing in biomolecular NMR. J. Biomol. NMR 1995, 6, 135–140. [Google Scholar] [CrossRef] [PubMed]
- Vranken, W.F.; Boucher, W.; Stevens, T.J.; Fogh, R.H.; Pajon, A.; Llinas, M.; Ulrich, E.L.; Markley, J.L.; Ionides, J.; Laue, E.D. The CCPN data model for NMR spectroscopy: Development of a software pipeline. Proteins 2005, 59, 687–696. [Google Scholar] [CrossRef] [PubMed]
- Andrushchenko, V.V.; Vogel, H.J.; Prenner, E.J. Optimization of the hydrochloric acid concentration used for trifluoroacetate removal from synthetic peptides. J. Pept. Soc. 2007, 13, 37–43. [Google Scholar] [CrossRef] [PubMed]





© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oppong, E.; Stier, G.; Gaal, M.; Seeger, R.; Stoeck, M.; Delsuc, M.-A.; Cato, A.C.B.; Kieffer, B. An Amyloidogenic Sequence at the N-Terminus of the Androgen Receptor Impacts Polyglutamine Aggregation. Biomolecules 2017, 7, 44. https://doi.org/10.3390/biom7020044
Oppong E, Stier G, Gaal M, Seeger R, Stoeck M, Delsuc M-A, Cato ACB, Kieffer B. An Amyloidogenic Sequence at the N-Terminus of the Androgen Receptor Impacts Polyglutamine Aggregation. Biomolecules. 2017; 7(2):44. https://doi.org/10.3390/biom7020044
Chicago/Turabian StyleOppong, Emmanuel, Gunter Stier, Miriam Gaal, Rebecca Seeger, Melanie Stoeck, Marc-André Delsuc, Andrew C. B. Cato, and Bruno Kieffer. 2017. "An Amyloidogenic Sequence at the N-Terminus of the Androgen Receptor Impacts Polyglutamine Aggregation" Biomolecules 7, no. 2: 44. https://doi.org/10.3390/biom7020044
APA StyleOppong, E., Stier, G., Gaal, M., Seeger, R., Stoeck, M., Delsuc, M. -A., Cato, A. C. B., & Kieffer, B. (2017). An Amyloidogenic Sequence at the N-Terminus of the Androgen Receptor Impacts Polyglutamine Aggregation. Biomolecules, 7(2), 44. https://doi.org/10.3390/biom7020044