Phytochemical Analysis and Evaluation of Antioxidant and Biological Activities of Extracts from Three Clauseneae Plants in Northern Thailand
Abstract
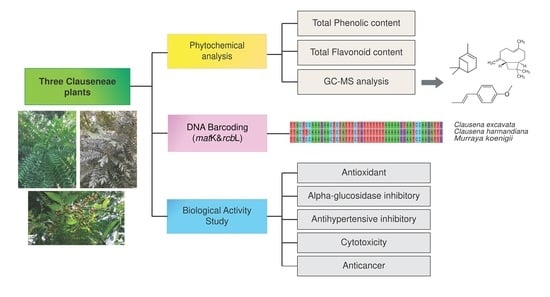
:1. Introduction
2. Results and Discussion
2.1. Genetic Variation Analysis
2.2. Phytochemical Analysis of Essential Oils
2.3. Total Phenolic and Total Flavonoid Contents
2.4. Determination of Methanol Extract and Essential Oil of Clausena excavata, Clausena harmandiana and Murraya koenigii against Human Normal Cells and Human Lung Cancer Cells
3. Materials and Methods
3.1. Plant Materials and Chemicals
3.2. Collection of Plant Material for Genetic Variation Analysis
3.2.1. Plant Sample Collection
3.2.2. DNA Extraction
3.2.3. PCR Amplication
3.2.4. Alignment of Sequences and Phylogenetic Analysis
3.3. Preparation of Plant Extracts
3.4. Phytochemical Analysis of Essential Oils
3.5. Determination of Total Flavonoid Contents
3.6. Determination of Total Phenolic Contents
3.7. Determination of Antioxidant Activities
3.7.1. DPPH Free Radical Scavenging Assay
- Percent inhibition = (Ao- As/Ao) × 100
- Where Ao is the absorbance of the control (containing all reagents except the test compound), and As is the absorbance of the mixture containing the test compound. The test sample concentrations providing 50% inhibition (IC50) were calculated from the plot of inhibition percentage against extract concentration values. The radical scavenging ability was presented IC50 values. The data were presented as the average of the triplicate analyses.
3.7.2. ABTS Radical Scavenging Activity
3.7.3. Ferric Reducing Antioxidant Power (FRAP) Assay
3.8. Determination of α-Glucosidase Inhibitory Activity
- Percent inhibition = (Ao − As/Ao) × 100
- Where Ao is the absorbance of the control, and As is the absorbance of the mixture containing the test compound. The data were presented as the average of the triplicate analyses.
3.9. Determination of Antihypertensive Activity
- Percent inhibition = [1 − (As/Ao)] × 100
- Where Ao is the absorbance of the control (containing all reagents except the test compound), and As is the absorbance of the mixture containing the test compound. The results of all experiments were expressed as mean ± standard deviation.
3.10. Antitumor Activity and Cell Toxicity Assay
3.11. Ethical Considerations
3.12. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pizzino, G.; Irrera, N.; Cucinotta, M.; Pallio, G.; Mannino, F.; Arcoraci, V.; Squadrito, F.; Altavilla, D.; Bitto, A. Oxidative stress: Harms and benefits for human health. Oxid. Med. Cell Longev. 2017, 2017, 8416763. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.C.; Lin, K.H.; Huang, M.Y.; Yang, C.M.; Shih, T.H.; Hsiung, T.C.; Lin, Y.C.; Tsao, F.C. Antioxidant activities of the methanol extracts of various parts of Phalaenopsis orchids with white, yellow, and purple flowers. Not. Bot. Horti Agrobo. Cluj Napoca. 2018, 46, 457–465. [Google Scholar] [CrossRef] [Green Version]
- Yen, G.C.; Chen, C.S.; Chang, W.T.; Wu, M.F.; Cheng, F.T.; Shiau, D.K.; Hsu, C.L. Antioxidant activity and anticancer effect of ethanolic and aqueous extracts of the roots of Ficus beecheyana and their phenolic components. J. Food Drug Anal. 2018, 26, 182–192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biparva, P.; Ehsani, M.; Hadjmohammadi, M.R. Dispersive liquid–liquid microextraction using extraction solvents lighter than water combined with high performance liquid chromatography for determination of synthetic antioxidants in fruit juice sample. J. Food Compost. Anal. 2012, 27, 87–94. [Google Scholar] [CrossRef]
- Sitthitharworn, W.; Wirasathien, L.; Kampiranont, L.; Khomdej, P.; Waiyasilp, W. Identification of Murraya koenigii (L.) Spreng using DNA barcoding technique based on the ITS sequence. Thai J. Pharm. Sci. 2010, 5, 202–205. [Google Scholar]
- Arbab, I.A.; Abdul, A.B.; Aspollah, M.; Abdullh, R.; Abdelwahab, S.I.; Ibrahim, M.Y.; Ali, L.Z. A review of traditional uses, phytochemical and pharmacological aspects of selected members of Clausena genus (Rutaceae). J. Med. Plant Res. 2012, 6, 5107–5118. [Google Scholar]
- Liu, J.; Li, C.J.; Du, Y.Q.; Li, L.; Sun, H.; Chen, N.H.; Zhang, D.M. Bioactive compounds from the stems of Clausena lansium. Molecules 2017, 22, 2226. [Google Scholar] [CrossRef] [Green Version]
- Palombo, E.A. Traditional medicinal plant extracts and natural products with activity against oral bacteria: Potential application in the prevention and treatment of oral diseases. Evid Based Complement Alternat Med. 2011, 2011, 680354. [Google Scholar] [CrossRef] [Green Version]
- Peng, W.; Fu, X.; Li, Y.; Xiong, Z.; Shi, Z.; Shi, X.; Zhang, F.; Huo, G.; Li, B. Phytochemical study of stem and leaf of Clausena lansium. Molecules 2019, 24, 3124. [Google Scholar] [CrossRef] [Green Version]
- Yenjai, C.; Sripontan, S.; Sriprajun, P.; Kittakoop, P.; Jintasirikul, A.; Tanticharoen, M.; Thebtaranonth, Y. Coumarins and carbazoles with antiplasmodial activity from Clausena harmandiana. Planta Med. 2000, 66, 277–279. [Google Scholar] [CrossRef] [Green Version]
- Wongthet, N.; Sanevas, N.; Schinnerl, J.; Valant-Vetschera, K.; Bacher, M.; Vajrodaya, S. Chemodiversity of Clausena excavata (Rutaceae) and related species: Coumarins and carbazoles. Biochem. Syst. 2018, 80, 84–90. [Google Scholar] [CrossRef]
- Mou, F.J.; Tu, T.Y.; Chen, Y.Z.; Zhang, D.X. Phylogenetic relationship of Clauseneae (Rutaceae) inferred from plastid and nuclear DNA data and taxonomic implication for some major taxa. Nord. J. Bot. 2018, 36, e01552. [Google Scholar] [CrossRef]
- Rao, B.R.; Rajput, D.K.; Mallavarapu, G.R. Chemical diversity in curry leaf (Murraya koenigii) essential oils. Food Chem. 2011, 126, 989–994. [Google Scholar]
- Balakrishnan, R.; Vijayraja, D.; Jo, S.H.; Ganesan, P.; Su-Kim, I.; Choi, D.K. Medicinal profile, phytochemistry, and pharmacological activities of Murraya koenigii and its primary bioactive compounds. Antioxidants 2020, 24, 101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ajay, S.; Rahul, S.; Sumit, G.; Paras, M.; Mishra, A.; Gaurav, A. Comprehensive review: Murraya koenigii Linn. Asian J. Pharm. Life Sci. 2011, 1, 417–425. [Google Scholar]
- Hebert, P.D.N.; Cywinska, A.; Ball, S.L.; de Waard, J.R. Biological Identifications through DNA Barcodes. Proc. Royal Soc. B Biol. Sci. 2003, 270, 313–321. [Google Scholar] [CrossRef] [Green Version]
- Shivakumar, V.S.; Appelhans, M.S.; Johnson, G.; Carlsen, M.; Zimmer, E.A. Analysis of whole chloroplast genomes from the genera of the Clauseneae, the curry tribe (Rutaceae, Citrus family). Mol. Phylogenet. Evol. 2017, 117, 135–140. [Google Scholar] [CrossRef]
- Hollingsworth, P.M.; Graham, S.W.; Little, D.P. Choosing and using a plant DNA barcode. PLoS ONE 2011, 6, e19254. [Google Scholar] [CrossRef]
- Penjor, T.; Yamamoto, M.; Uehara, M.; Ide, M.; Matsumoto, N.; Matsumoto, R.; Nagano, Y. Phylogenetic relationships of Citrus and its relatives based on matK gene sequences. PLoS ONE 2013, 8, e62574. [Google Scholar] [CrossRef] [Green Version]
- Duan, H.; Wang, W.; Zeng, Y.; Guo, M.; Zhou, Y. The screening and identification of DNA barcode sequences for Rehmannia. Sci. Rep. 2019, 9, 17295. [Google Scholar] [CrossRef] [Green Version]
- Gardner, S.; Sidisunthorn, P.; Chayamarit, K. Rutaceae. In Forest Trees of Southern Thailand; Amarin Printing and Publishing Plc.: Bangkok, Thailand, 2018; Volume 3 (Mo-Z), pp. 1968–1969. [Google Scholar]
- Li, D.Z.; Gao, L.M.; Li, H.T.; Wang, H.; Ge, X.J.; Liu, J.Q.; Chen, Z.D.; Zhou, S.L.; Chen, S.L.; Yang, J.B.; et al. Comparative analysis of a large dataset indicates that internal transcribed spacer (ITS) should be incorporated into the core barcode for seed plants. Proc. Nat. Acad. Sci. USA 2011, 108, 19641–19646. [Google Scholar] [PubMed] [Green Version]
- Cheng, S.S.; Chang, H.T.; Lin, C.Y.; Chen, P.S.; Huang, C.G.; Chen, W.J.; Chang, S.T. Insecticidal activities of leaf and twig essential oils from Clausena excavata against Aedes aegypti and Aedes albopictus larvae. Pest Manag. Sci. 2009, 65, 339–343. [Google Scholar] [CrossRef] [PubMed]
- Trung, H.D.; Thang, T.D.; Ban, P.H.; Hoi, T.M.; Dai, D.N.; Ogunwande, I.A. Terpene constituents of the leaves of five Vietnamese species of Clausena (Rutaceae). Nat. Prod. Res. 2014, 28, 622–630. [Google Scholar] [CrossRef] [PubMed]
- Songsiang, U.; Thongthoom, T.; Zeekpudsa, P.; Kukongviriyapan, V.; Boonyarat, C.; Wangboonskul, J.; Yenjai, C. Antioxidant activity and cytotoxicity against cholangiocarcinoma of carbazoles and coumarins from Clausena harmandiana. Sci. Asi. 2012, 38, 75–81. [Google Scholar] [CrossRef]
- Greger, H. Phytocarbazoles: Alkaloids with great structural diversity and pro-nounced biological activities. Phytochem. Rev. 2017, 16, 1095–1153. [Google Scholar] [CrossRef]
- Sukkaew, S.; Pripdeevech, P.; Thongpoon, C.; Machan, T.; Wongchuphan, R. Volatile constituents of Murraya koenigii fresh leaves using headspace solid phase microextraction—Gas chromatography—Mass spectrometry. Nat. Prod. Commun. 2014, 9, 1783–1786. [Google Scholar] [CrossRef] [Green Version]
- Nagappana, T.; Ramasamyb, P.; Vairappana, C.S. Chemotaxonomical markers in essential oil of Murraya koenigi. Nat. Prod. Commun. 2012, 7, 1375–1378. [Google Scholar]
- Chowdhury, J.U.; Bhuiyan, M.N.I.; Yusuf, M. Chemical composition of the leaf essential oils of Murraya koenigii (L.) Spreng and Murraya paniculata (L.) Jack. Bangladesh J. Pharmacol. 2008, 3, 59–63. [Google Scholar] [CrossRef] [Green Version]
- Verma, R.S.; Padalia, R.C.; Arya, V.; Chauhan, A. Aroma profiles of the curry leaf, Murraya koenigii (L.) Spreng. Chemotypes: Variability in north India during the year. Ind. Crops Prod. 2012, 36, 343–348. [Google Scholar] [CrossRef]
- Rajendran, M.P.; Pallaiyan, B.B.; Selvaraj, N. Chemical composition, antibacterial and antioxidant profile of essential oil from Murraya koenigii (L.) leaves. Avicenna J. Phytomed. 2014, 4, 200–214. [Google Scholar]
- Tripathi, Y.C.; Anjum, N.; Rana, A. Chemical composition and in vitro antifungal and antioxidant activities of essential oil from Murraya koenigii (L.) Spreng. Leaves. Asian J. Biomed. Pharm. 2018, 8, 6–13. [Google Scholar] [CrossRef] [Green Version]
- Gupta, S.; Paarakh, P.M.; Gavani, U. Isolation of phytoconstituents from the leaves of Murraya koenigii Linn. J. Pharm Res. 2009, 2, 1313–1314. [Google Scholar]
- Soares, J.R.; Dins, T.C.P.; Cunha, A.P.; Almeida, L.M. Antioxidant activity of some extracts of Thymus zygis. Free Radic. Res. 1997, 26, 469–478. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.G.; Xu, K.; Sang, Z.P.; Wei, R.R.; Liu, W.M.; Su, Y.L.; Yang, J.B.; Wang, A.G.; Ji, T.F.; Li, L.J. Alkenes with antioxidative activities from Murraya koenigii (L.) Spreng. Bioorganic Med. Chem. Lett. 2016, 26, 799–803. [Google Scholar] [CrossRef]
- Truong, D.H.; Nguyen, D.H.; Ta, N.T.A.; Bui, A.V.; Do, T.H.; Nguyen, H.C. Evaluation of the use of different solvents for phytochemical constituents, antioxidants, and in vitro anti-inflammatory activities of Severinia buxifolia. J. Food Qual. 2019, 2019, 8178294. [Google Scholar] [CrossRef] [Green Version]
- Albaayit, S.F.A.; Abba, Y.; Abdullah, R.; Abdullah, N. Evaluation of antioxidant activity and acute toxicity of Clausena excavata leaves extract. Evid. Based Complementary Altern. Med. 2014, 2014, 975450. [Google Scholar] [CrossRef] [Green Version]
- Thant, T.M.; Aminah, N.S.; Kristanti, A.N.; Ramadhan, R.; Aung, H.T.; Takaya, Y. Antidiabetes and antioxidant agents from Clausena excavata root as medicinal plant of Myanmar. Open Chem. 2019, 17, 1339–1344. [Google Scholar] [CrossRef] [Green Version]
- Arulselvan, P.; Senthilkumar, G.P.; Sathish Kumar, D.; Subramanian, S. Antidiabetic effect of Murraya koenigii leaves on streptozotocin induced diabetic rats. Pharmazie 2006, 61, 874–877. [Google Scholar]
- Yankuzo, H.; Ahmed, Q.U.; Santosa, R.I.; Akter, S.F.U.; Talib, N.A. Beneficial effect of the leaves of Murraya koenigii L. Spreng (Rutaceae) on diabetes-induced renal damage in vivo. J. Ethnopharmacol. 2011, 135, 88–94. [Google Scholar] [CrossRef]
- Nagarani, G.; Abirami, A.; Siddhuraju, P. A comparative study on antioxidant potentials, inhibitory activities against key enzymes related to metabolic syndrome, and anti-inflammatory activity of leaf extract from different Momordica species. Food Sci. Hum. Well 2014, 3, 36–46. [Google Scholar] [CrossRef] [Green Version]
- Sharifi, N.; Souri, E.; Ziai, S.A.; Amin, G.; Amanlou, M. Discovery of new angiotensin converting enzyme (ACE) inhibitors from medicinal plants to treat hypertension using an in vitro assay. DARU J. Pharm. Sci. 2013, 21, 74. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Feng, Z.L.; Wang, Y.T.; Lin, L.G. Anticancer carbazole alkaloids and coumarins from Clausena plants. A review. Chin. J. Nat. Med. 2017, 15, 881–888. [Google Scholar] [CrossRef]
- Tanruean, K.; Suwannarach, N.; Choonpicharn, S.; Lumyong, S. Evaluation of phytochemical constituents and biological activities of leaves and stems of Marsdenia glabra Cost. Int. Food Res. J. 2017, 24, 2572–2579. [Google Scholar]
- Muthumani, P.; Venkatraman, S.; Ramseshu, K.; Meera, R.; Devi, P.; Kameswari, B.; Eswarapriya, B. Pharmacological studies of anticancer, anti-inflammatory activities of Murraya koenigii L. Spreng in experimental animals. J. Pharm. Sci. Res. 2009, 1, 131–141. [Google Scholar]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [Green Version]
- Hillis, D.M.; Bull, J.J. An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst. Biol. 1993, 42, 182–192. [Google Scholar] [CrossRef]
- Tanruean, K.; Kaewnarin, K.; Suwannarach, N.; Lumyong, S. Comparative evaluation of phytochemicals, and antidiabetic and antioxidant activities of Cuscuta reflexa grown on different hosts in northern Thailand. Nat. Prod. Commun. 2017, 12, 51–54. [Google Scholar] [CrossRef] [Green Version]
- Kaewnarin, K.; Niamsup, H.; Shank, L.; Rakariyatham, N. Antioxidant and antidiabetic activities of some edible and medicinal plants. Chiang Mai J. Sci. 2014, 41, 105–116. [Google Scholar]
- Zhishen, J.; Mengcheng, T.; Jianming, W. The determination of flavonoid contents in mulberry and their scavenging effects on superoxide radicals. Food Chem. 1999, 64, 555–559. [Google Scholar] [CrossRef]
- Kamarul Zaman, M.A.; Azzeme, A.M.; Ramle, I.K.; Normanshah, N.; Ramli, S.N.; Shaharuddin, N.A.; Ahmad, S.; Abdullah, S.N.A. Induction, multiplication, and evaluation of antioxidant activity of Polyalthia bullata callus, a woody medicinal plant. Plants 2020, 9, 1772. [Google Scholar] [CrossRef] [PubMed]
- Radulescu, C.; Buruleanu, L.C.; Nicolescu, C.M.; Olteanu, R.L.; Bumbac, M.; Holban, G.C.; Simal-Gandara, J. Phytochemical profiles, antioxidant and antibacterial activities of grape (Vitis vinifera L.) seeds and skin from organic and conventional vineyards. Plants 2020, 9, 1470. [Google Scholar] [CrossRef] [PubMed]
- Thitilertdecha, N.; Teerawutgulrag, A.; Rakariyatham, N. Antioxidant and antimicrobial activities of Nephelium lappacium L. extracts. LWT Food Sci. Technol. 2008, 41, 2029–2035. [Google Scholar] [CrossRef]
- Gülçın, İ.; Oktay, M.; Kıreçcı, E.; Küfrevıoǧlu, Ö.İ. Screening of antioxidant and antimicrobial activities of anise (Pimpinella anisum L.) seed extracts. Food Chem. 2003, 83, 371–382. [Google Scholar] [CrossRef]
- Re, R.; Pellegrini, N.; Proteggente, A.; Pannala, A.; Yang, M.; Rice- Evans, C. Antioxidant activity applying an improved ABTS radical cation decolorization assay. Radic. Biol. Med. 1999, 26, 1231–1237. [Google Scholar] [CrossRef]
- Li, Y.; Guo, C.; Yang, J.; Wei, J.; Xu, J.; Cheng, S. Evaluation of antioxidant properties of pomegranate peel extract in comparison with pomegranate pulp extract. Food Chem. 2006, 96, 254–260. [Google Scholar] [CrossRef]
- Oki, T.; Matsui, T.; Osajima, Y. Inhibitory effect of α-glucosidase inhibitors varies according to its origin. J. Agric. Food Chem. 1999, 47, 550–553. [Google Scholar] [CrossRef] [PubMed]
- Adisakwattana, S.; Chantarasinlapin, P.; Thammarat, H.; Yibchok-Anun, S. A series of cinnamic acid derivatives and their inhibitory activity on intestinal α-glucosidase. J. Enzyme Inhib. Med. Chem. 2009, 24, 1194–1200. [Google Scholar] [CrossRef]
- Park, P.J.; Je, J.Y.; Kim, S.K. Angiotensin I converting enzyme (ACE) inhibitory activity of hetero-chitooligosaccharides prepared from partially different deacetylated chitosans. J. Agric. Food Chem. 2003, 51, 4930–4934. [Google Scholar] [CrossRef]
- Wang, P.; Henning, S.M.; Heber, D. Limitations of MTT and MTS-based assays for measurement of antiproliferative activity of green tea polyphenols. PLoS ONE 2010, 5, e10202. [Google Scholar] [CrossRef]





| Rt (min) a | Compound b | % Composition | ||
|---|---|---|---|---|
| C. excavata | C. harmandiana | M. koenigii | ||
| 5.617 | α-Pinene | - | 1.05 | 12.23 |
| 5.943 | Camphene | - | 9.61 | 0.27 |
| 5.998 | Sabinene | - | - | 0.05 |
| 6.685 | β-Terpinene | - | 7.87 | - |
| 6.703 | β-Pinene | - | 0.88 | 0.46 |
| 6.952 | 6-Methyl- 5-hepten-2-one | - | - | 0.28 |
| 7.068 | β-Mycene | - | 3.69 | 0.65 |
| 7.513 | α-Phellandrene | - | - | 0.63 |
| 7.693 | 3-Carene | 0.76 | 2.41 | 0.33 |
| 8.172 | p-Cymene | - | 1.27 | 0.53 |
| 8.406 | D-Limonene | - | 7.07 | - |
| 8.342 | β-Phellandrene | - | - | 3.67 |
| 8.456 | Eucalyptol | - | 0.5 | - |
| 8.616 | (E)-Ocimene | - | 0.9 | - |
| 8.947 | α-Ocimene | - | 0.57 | 0.78 |
| 9.403 | γ-Terpinene | - | 3.57 | - |
| 9.705 | (Z)-β-Terpineol | - | 0.4 | - |
| 10.419 | Terpinolene | 0.25 | 1.38 | 0.85 |
| 10.895 | 3,7-dimethyl-1,6-Octadien-3-ol | - | - | 0.57 |
| 11.761 | trans-1-Methyl-4-(1-methylethyl)-2-cyclohexen-1-ol | - | 0.29 | - |
| 13.986 | 4-Terpinenol | - | 3.34 | 0.09 |
| 14.849 | Estragole | 1.23 | 1.68 | 0.38 |
| 18.805 | Anethole | 86.72 | 46.09 | 26.02 |
| 21.137 | α-Cubebene | - | - | 0.15 |
| 22.030 | Ylangene | - | - | 0.16 |
| 22.220 | Copaene | - | - | 0.31 |
| 22.400 | cis-β-Guaiene | - | - | 0.16 |
| 22.925 | β-Elemene | - | - | 0.41 |
| 23.067 | 6,10,11,11-Tetramethyl-tricyclo[6.3.0.1(2,3)]undec-7-ene | - | - | 0.23 |
| 23.598 | α-Gurjunene | - | - | 0.70 |
| 24.202 | Caryophyllene | - | 0.67 | 21.15 |
| 24.695 | Zingiberene | - | - | 1.29 |
| 24.817 | Spathulenol | - | - | 1.25 |
| 24.978 | Eudesma-4(14),11-diene | - | - | 0.55 |
| 25.434 | α-Caryophyllene | - | 0.17 | 3.92 |
| 26.739 | β-helmiscapene | - | - | 3.81 |
| 27.105 | γ-Elemene | - | 0.19 | - |
| 27.126 | α-Selinene | 0.34 | - | 6.10 |
| 30.285 | Spathulenol | 0.56 | - | - |
| 30.445 | Caryophyllene oxide | - | - | 0.85 |
| Total | 89.86 | 93.60 | 88.87 | |
| Plant Extracts | Percent Yield (%Yield) | Total Phenolic Content (mg GAE/g Extract) | Total Flavonoid Content (mg QE/g Extract) |
|---|---|---|---|
| C. excavata methanol extract | 5.52 ± 0.32a | 22.89 ± 0.93c | 30.89 ± 2.15d |
| C. excavata essential oil | 0.83 ± 0.06d | 9.70 ± 0.72d | 23.91 ± 1.98e |
| C. harmandiana methanol extract | 4.12 ± 0.19c | 19.71 ± 0.83c | 39.95 ± 0.63c |
| C. harmandiana essential oil | 0.64 ± 0.08d | 7.07 ± 0.73d | 16.82 ± 1.25f |
| M. koenigii methanol extract | 4.92 ± 0.39b | 43.50 ± 4.30a | 66.13 ± 1.69a |
| M. koenigii essential oil | 0.86 ± 0.09d | 36.28 ± 1.65b | 50.57 ± 1.11b |
| Plant Extracts | Antioxidant Activity | ||
|---|---|---|---|
| DPPH Free Radical Scavenging (IC50, ug/mL) | ABTS Cation Free Radical Scavenging (mg GAE/g extract) | Ferric Reducing Antioxidant Power (mg GAE/g extract) | |
| C. excavata methanol extract | 904.53 ± 3.23c | 88.65 ± 0.71b | 16.48 ± 0.72d |
| C. excavata essential oil | 2059.29 ± 83.13d | 12.27 ± 0.1.75d | 5.07 ± 0.17e |
| C. harmandiana methanol extract | 2037.66 ± 39.23d | 80.11 ± 1.01c | 19.07 ± 0.55c |
| C. harmandiana essential oil | 2865.26 ± 8.22e | 12.83 ± 1.04d | 5.37 ± 0.32e |
| M. koenigii methanol extract | 95.54 ± 3.46a | 118.12 ± 1.01a | 48.15 ± 1.21a |
| M. koenigii essential oil | 167.74 ± 6.97b | 79.52 ± 1.01c | 28.22 ± 0.94b |
| Plant Extracts | α-Glucosidase Inhibitory Activity (% Inhibition) | Antihypertensive Inhibitory Activity (% Inhibition) |
|---|---|---|
| C. excavata methanol extract | 49.30 ± 1.10c | 47.63 ± 1.11c |
| C. excavata essential oil | 36.20 ± 1.49e | 38.90 ± 1.05d |
| C. harmandiana methanol extract | 41.42 ± 0.65d | 58.10 ± 1.75b |
| C. harmandiana essential oil | 24.33 ± 0.91f | 31.07 ± 1.16e |
| M. koenigii methanol extract | 84.55 ± 0.49a | 84.95 ± 1.24a |
| M. koenigii essential oil | 52.39 ± 1.16b | 39.33 ± 1.13d |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tanruean, K.; Poolprasert, P.; Suwannarach, N.; Kumla, J.; Lumyong, S. Phytochemical Analysis and Evaluation of Antioxidant and Biological Activities of Extracts from Three Clauseneae Plants in Northern Thailand. Plants 2021, 10, 117. https://doi.org/10.3390/plants10010117
Tanruean K, Poolprasert P, Suwannarach N, Kumla J, Lumyong S. Phytochemical Analysis and Evaluation of Antioxidant and Biological Activities of Extracts from Three Clauseneae Plants in Northern Thailand. Plants. 2021; 10(1):117. https://doi.org/10.3390/plants10010117
Chicago/Turabian StyleTanruean, Keerati, Pisit Poolprasert, Nakarin Suwannarach, Jaturong Kumla, and Saisamorn Lumyong. 2021. "Phytochemical Analysis and Evaluation of Antioxidant and Biological Activities of Extracts from Three Clauseneae Plants in Northern Thailand" Plants 10, no. 1: 117. https://doi.org/10.3390/plants10010117
APA StyleTanruean, K., Poolprasert, P., Suwannarach, N., Kumla, J., & Lumyong, S. (2021). Phytochemical Analysis and Evaluation of Antioxidant and Biological Activities of Extracts from Three Clauseneae Plants in Northern Thailand. Plants, 10(1), 117. https://doi.org/10.3390/plants10010117