The RNA Directed DNA Methylation (RdDM) Pathway Regulates Anthocyanin Biosynthesis in Crabapple (Malus cv. spp.) Leaves by Methylating the McCOP1 Promoter
Abstract
:1. Introduction
2. Results
2.1. There Is a Negative Correlation between McCOP1 Promoter Methylation Levels and Transcription in Crabapple Leaves
2.2. DNA Methylation Levels of the McCOP1 Promoter Change after Treatment with Methylation Inhibitors
2.3. AGO4 Recognize the SINE Site in the McCOP1 Promoter
2.4. McRDM1 Silencing Inhibits Anthocyanin Biosynthesis in Crabapple
2.5. AtRDM1 Contributes to Anthocyanin Accumulation in A. thaliana under Stress Conditions
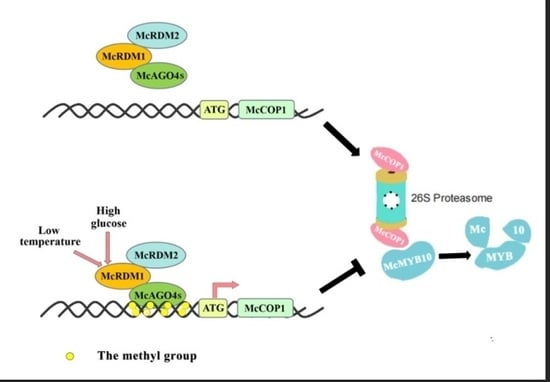
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth
4.2. Anthocyanin Quantification
4.3. Amplification of the McAGO4-A, McAGO4-Like, McDRM2 and McRDM1 and the McCOP1 Promoter
4.4. Measurement of Methylation Levels
4.5. Expression Analysis
4.6. Yeast One-Hybrid Assay
4.7. EMSA
4.8. BiFC
4.9. RDM1 Silencing in Crabapple Plantlets and Apple Fruit
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Treutter, D. Significance of flavonoids in plant resistance and enhancement of their biosynthesis. Plant Biol. 2005, 7, 581–591. [Google Scholar] [CrossRef]
- Misra, P.; Pandey, A.; Tiwari, M.; Chandrashekar, K.; Sidhu, O.P.; Asif, M.H.; Chakrabarty, D.; Singh, P.K. Modulation of transcriptome and metabolome of tobacco by Arabidopsis transcription factor, AtMYB12, leads to insect resistance. Plant Physiol. 2010, 152, 2258–2268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Onkokesung, N.; Reichelt, M.; van Doorn, A.; Schuurink, R.C.; van Loon, J.J.; Dicke, M. Modulation of flavonoid metabolites in Arabidopsis thaliana through overexpression of the MYB75 transcription factor: Role of kaempferol-3,7-dirhamnoside in resistance to the specialist insect herbivore Pieris brassicae. J. Exp. Bot. Adv. Access 2014, 65, 2203–2217. [Google Scholar] [CrossRef] [Green Version]
- Page, M.; Sultana, N.; Paszkiewicz, K.; Florance, H.; Smirnoff, N. The influence of ascorbate on anthocyanin accumulation during high light acclimation in Arabidopsis thaliana: Further evidence for redox control of anthocyanin synthesis. Plant Cell Environ. 2012, 35, 388–404. [Google Scholar] [CrossRef]
- Medic, N.; Tramer, F.; Passamonti, S. Anthocyanins in colorectal cancer prevention. A systematic review of the literature in search of molecular oncotargets. Front. Pharmacol. 2019, 10, 675. [Google Scholar] [CrossRef]
- Cremonini, E.; Daveri, E.; Mastaloudis, A.; Adamo, A.M.; Mills, D.; Kalanetra, K.; Hester, S.N.; Wood, S.M.; Fraga, C.G.; Oteiza, P.I. Anthocyanins protect the gastrointestinal tract from high fat diet-induced alterations in redox signaling, barrier integrity and dysbiosis. Redox Biol. 2019, 26, 101269. [Google Scholar] [CrossRef]
- Mehrtens, F.; Kranz, H.; Bednarek, P.; Weisshaar, B. The Arabidopsis transcription factor MYB12 is a flavonol-specific regulator of phenylpropanoid biosynthesis. Plant Physiol. 2005, 138, 1083–1096. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stracke, R.; Ishihara, H.; Huep, G.; Barsch, A.; Mehrtens, F.; Niehaus, K.; Weisshaar, B. Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J. 2007, 50, 660–677. [Google Scholar] [CrossRef] [Green Version]
- Allan, A.C.; Hellen, R.P.; Laing, W.A. MYB transcription factors that colour our fruit. Trends Plant Sci. 2008, 13, 99–102. [Google Scholar] [CrossRef]
- Gu, Z.Y.; Zhu, J.; Hao, Q.; Yuan, Y.W.; Duan, Y.W.; Men, S.Q.; Wang, Q.Y.; Hou, Q.Z.; Liu, Z.A.; Shu, Q.Y.; et al. A novel R2R3-MYB transcription factor contributes to petal blotch formation by regulating organ-specific expression of PsCHS in tree peony (Paeonia suffruticosa). Plant Cell Physiol. 2019, 60, 599–611. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.H.; Tang, W.; Hu, Y.W.; Zhang, Y.B.; Sun, J.Q.; Guo, X.L.; Lu, H.; Yang, Y.; Fang, C.B.; Niu, X.L.; et al. A MYB/bHLH complex regulates tissue-specific anthocyanin biosynthesis in the inner pericarp of red-centered kiwifruit Actinidia chinensis cv. Hongyang. Plant J. 2019, 99, 359–378. [Google Scholar] [CrossRef]
- Xu, Z.S.; Yang, Q.Q.; Feng, K.; Yu, X.; Xiong, A.S. DcMYB113, a root-specific R2R3-MYB, conditions anthocyanin biosynthesis and modification in carrot. Plant Biotechnol. J. 2020, 18, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Takos, A.M.; Jaffé, F.W.; Jacob, S.R.; Bogs, J.; Robinson, S.P.; Walker, A.R. Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol. 2006, 9, 1216–1232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Espley, R.V.; Hellens, R.P.; Putterill, J.; Stevenson, D.E.; Kutty-Amma, S.; Allan, A.C. Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J. 2007, 49, 414–427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ban, Y.; Honda, C.; Hatsuyama, Y.; Igarashi, M.; Bessho, H.; Moriguchi, T. Isolation and functional analysis of a MYB transcription factor gene that is a key regulator for the development of red coloration in apple skin. Plant Cell Physiol. 2007, 48, 958–970. [Google Scholar] [CrossRef]
- Chagné, D.; Lin-Wang, K.; Espley, R.V.; Volz, R.K.; How, N.M.; Rouse, S.; Brendolise, C.; Carlisle, C.M.; Kumar, S.; De Silva, N.; et al. An ancient duplication of apple MYB transcription factors is responsible for novel red fruit-flesh phenotypes. Plant Physiol. 2013, 161, 225–239. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.Y.; Mao, K.; Zhao, C.; Zhao, X.Y.; Zhang, H.L.; Shu, H.R.; Hao, Y.J. MdCOP1 ubiquitin E3 ligases interact with MdMYB1 to regulate light-induced anthocyanin biosynthesis and red fruit coloration in apple. Plant Physiol. 2012, 160, 1011–1022. [Google Scholar] [CrossRef] [Green Version]
- Wu, M.; Si, M.; Li, X.Y.; Song, L.Y.; Liu, J.L.; Zhai, R.; Cong, L.; Yue, R.R.; Yang, C.Q.; Ma, F.W.; et al. PbCOP1.1 contributes to the negative regulation of anthocyanin biosynthesis in pear. Plants 2019, 8, 39. [Google Scholar] [CrossRef] [Green Version]
- Peng, T.; Saito, T.; Honda, C.; Ban, Y.; Kondo, S.; Liu, J.H.; Hatsuyama, Y.; Moriguchi, T. Screening of UV-B-induced genes from apple peels by SSH: Possible involvement of MdCOP1-mediated signaling cascade genes in anthocyanin accumulation. Physiol. Plant 2013, 148, 432–444. [Google Scholar] [CrossRef]
- Law, J.A.; Jacobsen, S.E. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 2010, 11, 204–220. [Google Scholar] [CrossRef]
- Mbichi, R.W.; Wang, Q.F.; Wan, T. RNA directed DNA methylation and seed plant genome evolution. Plant Cell Rep. 2020, 39, 983–996. [Google Scholar] [CrossRef]
- Gao, Z.H.; Liu, H.L.; Daxinger, L.; Pontes, O.; He, X.J.; Qian, W.Q.; Lin, H.X.; Xie, M.T.; Lorkovic, Z.J.; Zhang, S.D.; et al. An RNA polymerase II- and AGO4-associated protein acts in RNA-directed DNA methylation. Nature 2010, 465, 106–109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Au, P.C.K.; Dennis, E.S.; Wang, M.B. Analysis of Argonaute 4-Associated Long Non-Coding RNA in Arabidopsis thaliana Sheds Novel Insights into Gene Regulation through RNA-Directed DNA Methylation. Genes 2017, 8, 198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, F.; Axtell, M.J. AGO4 is specifically required for heterochromatic siRNA accumulation at Pol V-dependent loci in Arabidopsis thaliana. Plant J. 2017, 90, 37–47. [Google Scholar] [CrossRef] [Green Version]
- Qi, Y.; He, X.; Wang, X.J.; Kohany, O.; Jurka, J.; Hannon, G.J. Distinct catalytic and non-catalytic roles of ARGONAUTE4 in RNA-directed DNA methylation. Nature 2006, 443, 1008–1012. [Google Scholar] [CrossRef]
- Dinh, T.T.; O’Leary, M.; Won, S.Y.; Li, S.; Arroyo, L.; Liu, X.; Defries, A.; Zheng, B.; Cutler, S.R.; Chen, X. Generation of a luciferase-based reporter for CHH and CG DNA methylation in Arabidopsis thaliana. Silence 2013, 4, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Ingouff, M.; Selles, B.; Michaud, C.; Vu, T.M.; Berger, F.; Schorn, A.J.; Autran, D.; Van Durme, M.; Nowack, M.K.; Martienssen, R.A.; et al. Live-cell analysis of DNA methylation during sexual reproduction in Arabidopsis reveals context and sex-specific dynamics controlled by noncanonical RdDM. Genes Dev. 2017, 31, 72–83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, Q.; Rowley, M.J.; Böhmdorfer, G.; Sandhu, D.; Gregory, B.D.; Wierzbicki, A.T. RNA polymerase V targets transcriptional silencing components to promoters of protein-coding genes. Plant J. 2013, 73, 179–189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, P.H.; Wittmeyer, K.T.; Lee, T.F.; Meyers, B.C.; Chopra, S. Overlapping RdDM and non-RdDM mechanisms work together to maintain somatic repression of a paramutagenic epiallele of maize pericarp color1. PLoS ONE 2017, 12, e0187157. [Google Scholar] [CrossRef] [Green Version]
- Cheng, J.; Niu, Q.; Zhang, B.; Chen, K.; Yang, R.; Zhu, J.K.; Zhang, Y.; Lang, Z. Downregulation of RdDM during strawberry fruit ripening. Genome Biol. 2018, 19, 1–14. [Google Scholar] [CrossRef]
- Vaucheret, H. Plant ARGONAUTES. Trends Plant Sci. 2008, 13, 350–358. [Google Scholar] [CrossRef]
- Iwasaki, M.; Hyvärinen, L.; Piskurewicz, U.; Lopez-Molina, L. Non-canonical RNA-directed DNA methylation participates in maternal and environmental control of seed dormancy. eLife 2019, 8, e37434. [Google Scholar] [CrossRef]
- Kirkbride, R.C.; Lu, J.; Zhang, C.; Mosher, R.A.; Baulcombe, D.C.; Chen, Z.J. Maternal small RNAs mediate spatial-temporal regulation of gene expression, imprinting, and seed development in Arabidopsis. Proc. Natl. Acad. Sci. USA 2019, 116, 2761–2766. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chow, H.T.; Chakraborty, T.; Mosher, R.A. RNA-directed DNA Methylation and sexual reproduction: Expanding beyond the seed. Curr. Opin. Plant Biol. 2020, 54, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.H.; Wang, N.; Chen, M.; Zhang, R.; Sun, Q.G.; Xu, H.F.; Zhang, Z.Y.; Wang, Y.C.; Sui, X.Q.; Wang, S.F.; et al. Methylation of MdMYB1 locus mediated by RdDM pathway regulates anthocyanin biosynthesis in apple. Plant Biotechnol. J. 2020, 18, 1736–1748. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zilberman, D.; Cao, X.; Jacobsen, S.E. ARGONAUTE4 Control of Locus-Specific siRNA Accumulation and DNA and Histone Methylation. Science 2003, 299, 716–719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lahmy, S.; Pontier, D.; Bies-Etheve, N.; Laudié, M.; Feng, S.; Jobet, E.; Hale, C.J.; Cooke, R.; Hakimi, M.A. Evidence for ARGONAUTE4-DNA interactions in RNA-directed DNA methylation in plants. Genes Dev. 2016, 30, 2565–2570. [Google Scholar] [CrossRef]
- Li, P.; Li, Y.; Zhang, F.; Zhang, G.; Jiang, X.; Yu, H.; Hou, B. The Arabidopsis UDP glycosyltransferases UGT79B2 and UGT79B3, contribute to cold, salt and drought stress tolerance via modulating anthocyanin accumulation. Plant J. 2017, 89, 85–103. [Google Scholar] [CrossRef] [Green Version]
- Liang, J.; He, J.X. Protective role of anthocyanins in plants under low nitrogen stress. Biochem. Biophys. Res. Commun. 2018, 498, 946–953. [Google Scholar] [CrossRef]
- Naydenov, M.; Baev, V.; Apostolova, E.; Gospodinova, N.; Sablok, G.; Gozmanova, M.; Yahubyan, G. High-temperature effect on genes engaged in DNA methylation and affected by DNA methylation in Arabidopsis. Plant Physiol. Biochem. 2015, 87, 102–108. [Google Scholar] [CrossRef]
- Kvaalen, H.; Johnsen, Ø. Timing of bud set in Picea abies is regulated by a memory of temperature during zygotic and somatic embryogenesis. New Phytol. 2008, 177, 49–59. [Google Scholar] [CrossRef]
- Liu, X.J.; Chuang, Y.N.; Chiou, C.Y.; Chin, D.C.; Shen, F.Q.; Yeh, K.W. Methylation effect on chalcone synthase gene expression determines anthocyanin pigmentation in floral tissues of two Oncidium orchid cultivars. Planta 2012, 236, 401–409. [Google Scholar] [CrossRef]
- Wang, Z.G.; Meng, D.; Wang, A.D.; Li, T.L.; Jiang, S.L.; Cong, P.H.; Li, T.Z. The methylation of the PcMYB10 promoter is associated with green-skinned sport in Max Red Bartlett pear. Plant J. 2013, 162, 885–896. [Google Scholar] [CrossRef] [Green Version]
- Wan, H.; Zhang, J.; Song, T.; Tian, J.; Yao, Y. Promotion of flavonoid biosynthesis in leaves and calli of ornamental crabapple (Malus sp.) by high carbon to nitrogen ratios. Front. Plant Sci. 2015, 6, 673. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brendolise, C.; Espley, R.V.; Lin, W.K.; Laing, W.; Peng, Y.; McGhie, T.; Dejnoprat, S.; Tomes, S.; Hellens, R.P.; Allan, A.C. Multiple copies of a simple MYB-binding site confers trans-regulation by specific flavonoid-related R2R3 MYBs in diverse species. Front. Plant Sci. 2017, 8, 1864. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, K.T.; Zhang, J.; Kang, Y.H.; Chen, M.C.; Song, T.T.; Geng, H.; Tian, J.; Yao, Y.C. McMYB10 modulates the expression of a Ubiquitin Ligase, McCOP1 during leaf coloration in crabapple. Front. Plant Sci. 2018, 9, 704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wongpalee, S.P.; Liu, S.; Gallego-Bartolomé, J.; Leitner, A.; Aebersold, R.; Liu, W.; Yen, L.; Nohales, M.A.; Kuo, P.H.; Vashisht, A.A.; et al. CryoEM structures of Arabidopsis DDR complexes involved in RNA-directed DNA methylation. Nat. Commun. 2019, 10, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Sasaki, T.; Lorković, Z.J.; Liang, S.C.; Matzke, A.J.; Matzke, M. The ability to form homodimers is essential for RDM1 to function in RNA-directed DNA methylation. PLoS ONE 2014, 9, e88190. [Google Scholar] [CrossRef] [Green Version]
- Baulcombe, D. RNA silencing in plants. Nature 2004, 431, 356–363. [Google Scholar] [CrossRef]
- Chan, S.W.; Henderson, I.R.; Jacobsen, S.E. Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat. Rev. Genet. 2005, 6, 351–360. [Google Scholar] [CrossRef] [PubMed]
- Matzke, M.; Matzke, A.J.; Kanno, T.; Huettel, B.; Mette, M.F.; Aufsatz, W.; Jaligot, E.; Daxinger, L.; Kreil, D.P. Atypical RNA polymerase subunits required for RNA-directed DNA methylation. Nat. Genet. 2005, 37, 761–765. [Google Scholar] [CrossRef]
- Matzke, M.; Kanno, T.; Daxinger, L.; Huettel, B.; Matzke, A.J.M. RNA-mediated chromatin-based silencing in plants. Curr. Opin. Cell Biol. 2009, 21, 367–376. [Google Scholar] [CrossRef] [PubMed]
- Havecker, E.R.; Wallbridge, L.M.; Hardcastle, T.J.; Bush, M.S.; Kelly, K.A.; Dunn, R.M.; Schwach, F.; Doonan, J.H.; Baulcombe, D.C. The Arabidopsis RNA-directed DNA methylation argonautes functionally diverge based on their expression and interaction with target loci. Plant Cell 2010, 22, 321–334. [Google Scholar] [CrossRef] [Green Version]
- Cao, X.; Jacobsen, S.E. Role of the Arabidopsis DRM methyltransferases in de novo DNA methylation and gene silencing. Curr. Biol. 2002, 12, 1138–1144. [Google Scholar] [CrossRef] [Green Version]
- Cao, X.; Jacobsen, S.E. Locus-specific control of asymmetric and CpNpG methylation by the DRM and CMT3 methyltransferase genes. Proc. Natl. Acad. Sci. USA 2002, 99, 16491–16498. [Google Scholar] [CrossRef] [Green Version]
- Wierzbicki, A.T. The role of long non-coding RNA in transcriptional gene silencing. Curr. Opining Plant Biol. 2012, 15, 517–522. [Google Scholar] [CrossRef]
- Rowley, M.J.; Rothi, M.H.; Böhmdorfer, G.; Kuciński, J.; Wierzbicki, A.T. Long-range control of gene expression via RNA-directed DNA methylation. PLoS Genet. 2017, 13, e1006749. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, T.; Ma, H.Y.; Zhang, J.; Wu, T.; Song, T.T.; Tian, J.; Yao, Y.C. Systematic identification of long noncoding RNAs expressed during light induced anthocyanin accumulation in apple fruit. Plant J. 2019, 100, 572–590. [Google Scholar] [CrossRef]
- Song, T.T.; Li, K.T.; Wu, T.; Wang, Y.; Zhang, X.Z.; Xu, X.F.; Yao, Y.C.; Han, Z.H. Identification of new regulators through transcriptome analysis that regulate anthocyanin biosynthesis in apple leaves at low temperatures. PLoS ONE 2019, 14, e0210672. [Google Scholar]
- Zhang, Y.C.; Zhang, J.; Song, T.T.; Li, J.; Tian, J.; Jin, K.N.; Yao, Y.C. Low medium pH value enhances anthocyanin accumulation in Malus crabapple leaves. PLoS ONE 2014, 9, e97904. [Google Scholar] [CrossRef]
- An, J.P.; Wang, X.; Hao, Y.J. BTB/TAZ protein MdBT2 integrates multiple hormonal and environmental signals to regulate anthocyanin biosynthesis in apple. J. Integr. Plant Biol. 2020, 62, 1643–1646. [Google Scholar] [CrossRef]
- An, J.P.; Zhang, X.; Bi, S.; You, C.; Wang, X.; Hao, Y.J. The ERF transcription factor MdERF38 promotes drought stress-induced anthocyanin biosynthesis in apple. Plant J. 2019, 101, 573–589. [Google Scholar] [CrossRef]
- An, J.P.; Wang, X.F.; Zhang, X.W.; Xu, H.F.; Bi, S.Q.; You, C.X.; Hao, Y.J. An apple MYB transcription factor regulates cold tolerance and anthocyanin accumulation and undergoes MIEL1-mediated degradation. Plant Biotechnol. J. 2020, 18, 337–353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Movahedi, A.; Zhang, J.; Sun, W.; Mohammadi, K.; Almasi Zadeh Yaghuti, A.; Wei, H.; Wu, X.; Yin, T. Functional analyses of PtRDM1 gene overexpression in poplars and evaluation of its effect on DNA methylation and response to salt stress. Plant Physiol. Biochem. 2018, 127, 64–73. [Google Scholar] [CrossRef]
- Zhu, Y.C.; Zhang, B.; Allan, A.C.; Lin-Wang, K.; Zhao, Y.; Wang, K.; Chen, K.S.; Xu, C.J. DNA demethylation is involved in the regulation of temperature-dependent anthocyanin accumulation in peach. Plant J. 2020, 102, 965–976. [Google Scholar] [CrossRef]
- Tian, J.; Peng, Z.; Zhang, J.; Song, T.T.; Wan, H.H.; Zhang, M.; Yao, Y.C. McMYB10 regulates coloration via activating McF3′H and later structural genes in ever red leaf crabapple. Plant Biotechnol. J. 2015, 13, 1–14. [Google Scholar] [CrossRef]
- Chirinos, R.; Campos, D.; Betalleluz, I.; Giusti, M.M.; Schwartz, S.J.; Tian, Q.; Pedreschi, R.; Larondelle, Y. High-performance liquid chromatography with photodiode array detection (HPLC-DAD)/HPLC-mass spectrometry (MS) profiling of anthocyanins from Andean Mashua Tubers (Tropaeolum tuberosum Ruíz and Pavón) and their contribution to the overall antioxidant activity. J. Agric. Food Chem. 2006, 54, 7089–7097. [Google Scholar]
- Telias, A.; Lin-Wang, K.; Stevenson, D.E.; Cooney, J.M.; Hellens, R.P.; Allan, A.C.; Hoover, E.E.; Bradeen, J.M. Apple skin patterning is associated with differential expression of MYB10. BMC Plant Biol. 2011, 11, 93. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative Pcr and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Lin, Q.; Lan, J.; Zhang, T.; Wan, J. WRKY Transcription Factor OsWRKY29 Represses Seed Saitou, N.; Nei, M. The neighbor-joining method—A new method for reconstructing phylogenetic rrees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Zhao, S.L.; Liang, C.Y.; Zhang, W.J.; Tang, X.C.; Peng, H.Y. Characterization of the RNA-binding domain in the Dendrolimus punctatus cytoplasmic polyhedrosis virus nonstructural protein p44. Virus Res. 2005, 114, 80–88. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.H.; Guo, S.Y.; Xu, Y.Y.; Li, C.H.; Zhang, Z.Y.; Zhang, D.J.; Xu, S.J.; Zhang, C.; Chong, K. OsmiR396d-regulated OsGRFs function in floral organogenesis in rice through binding to their targets OsJMJ706 and OsCR4. Plant Physiol. 2014, 165, 160–174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Charity, J.A.; Holland, L.; Grace, L.J.; Walter, C. Consistent and stable expression of the nptII, uidA and bar genes in transgenic Pinus radiata after Agrobacterium tumefaciens-mediated transformation using nurse cultures. Plant Cell Rep. 2005, 23, 606–616. [Google Scholar] [CrossRef] [PubMed]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xing, Y.; Xie, Z.; Sun, W.; Sun, Y.; Han, Z.; Zhang, S.; Tian, J.; Zhang, J.; Yao, Y. The RNA Directed DNA Methylation (RdDM) Pathway Regulates Anthocyanin Biosynthesis in Crabapple (Malus cv. spp.) Leaves by Methylating the McCOP1 Promoter. Plants 2021, 10, 2466. https://doi.org/10.3390/plants10112466
Xing Y, Xie Z, Sun W, Sun Y, Han Z, Zhang S, Tian J, Zhang J, Yao Y. The RNA Directed DNA Methylation (RdDM) Pathway Regulates Anthocyanin Biosynthesis in Crabapple (Malus cv. spp.) Leaves by Methylating the McCOP1 Promoter. Plants. 2021; 10(11):2466. https://doi.org/10.3390/plants10112466
Chicago/Turabian StyleXing, Yifan, Ziyi Xie, Weilei Sun, Yuying Sun, Zhenyun Han, Shiya Zhang, Ji Tian, Jie Zhang, and Yuncong Yao. 2021. "The RNA Directed DNA Methylation (RdDM) Pathway Regulates Anthocyanin Biosynthesis in Crabapple (Malus cv. spp.) Leaves by Methylating the McCOP1 Promoter" Plants 10, no. 11: 2466. https://doi.org/10.3390/plants10112466
APA StyleXing, Y., Xie, Z., Sun, W., Sun, Y., Han, Z., Zhang, S., Tian, J., Zhang, J., & Yao, Y. (2021). The RNA Directed DNA Methylation (RdDM) Pathway Regulates Anthocyanin Biosynthesis in Crabapple (Malus cv. spp.) Leaves by Methylating the McCOP1 Promoter. Plants, 10(11), 2466. https://doi.org/10.3390/plants10112466