Quinoa (Chenopodium quinoa Willd.): Genetic Diversity According to ISSR and SCoT Markers, Relative Gene Expression, and Morpho-Physiological Variation under Salinity Stress
Abstract
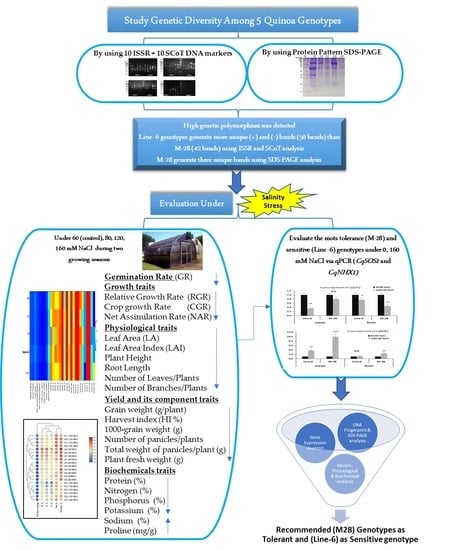
:1. Introduction
2. Materials and Methods
2.1. Germplasm, Experimental Setup, and Growth Conditions
2.2. Measurements of Growth and Developmental Parameters
2.2.1. Germination Rate
2.2.2. Morpho-Physiological Traits
2.2.3. Yield and Its Components
2.2.4. Measurements of Chemical Compositions
2.3. Molecular Characterization
2.3.1. ISSR and SCoT Marker Analysis
DNA Extraction and ISSR and SCot Amplification
2.3.2. Protein Analysis
2.3.3. Gene Expression Analysis
2.4. Statistical Analysis
3. Results
3.1. Genetic Diversity Analysis
3.1.1. ISSR and SCoT Marker Characterization
3.1.2. Protein Pattern (SDS-PAGE)
3.2. Overall Performance of Morpho-Physiological Traits under Salinity Stress
3.3. Biochemical Analysis under Salinity Stress
3.4. Gene Expression Analysis under Salinity Stress
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- González, J.A.; Hinojosa, L.; Mercado, M.I.; Fernández-Turiel, J.-L.; Bazile, D.; Ponessa, G.I.; Eisa, S.; González, D.A.; Rejas, M.; Hussin, S.; et al. A Long Journey of CICA-17 Quinoa Variety to Salinity Conditions in Egypt: Mineral Concentration in the Seeds. Plants 2021, 10, 407. [Google Scholar] [CrossRef] [PubMed]
- Bazile, D.; Jacobsen, S.E.; Verniau, A. The global expansion of quinoa: Trends and limits. Front. Plant Sci. 2016, 7, 622–629. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Pando, L.R.; Aguilar-Castellanos, E.; Ibañez-Tremolada, M. Quinoa (Chenopodium quinoa Willd.) breeding. In Advances in Plant Breeding Strategies: Cereals; Al-Khayri, J., Jain, S., Johnson, D., Eds.; Springer: Cham, Switzerland, 2019; pp. 259–316. [Google Scholar]
- Adolf, V.I.; Shabala, S.; Andersen, M.N.; Razzaghi, F.; Jacobsen, S.E. Varietal differences of quinoa’s tolerance to saline conditions. Plant Soil 2012, 357, 117–129. [Google Scholar] [CrossRef]
- Fuentes, F.; Bhargava, A. Morphological analysis of quinoa germplasm grown under lowland desert conditions. J. Agron. Crop Sci. 2011, 197, 124–134. [Google Scholar] [CrossRef]
- González, J.A.; Eisa, S.S.S.; Hussin, S.A.E.S.; Prado, F.E. Quinoa: An incan crop to face global changes in agriculture. In Quinoa: Improvement and Sustainable Production; Murphy, K., Matanguihan, J., Eds.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2015; pp. 1–18. ISBN 978-1-118-62804-1. [Google Scholar]
- Vilcacundo, R.; Hernández-Ledesma, B. Nutritional and biological value of quinoa (Chenopodium quinoa Willd.). Curr. Opin. Food Sci. 2017, 14, 1–6. [Google Scholar] [CrossRef]
- Gordillo-Bastidas, E.; Díaz-Rizzolo, D.A.; Roura, E.; Massanés, T.; Gomis, R. Quinoa (Chenopodium quinoa Willd), from Nutritional Value to Potential Health Benefits: An Integrative Review. J. Nutr. Food Sci. 2016, 6, 497. [Google Scholar] [CrossRef]
- Angeli, V.; Miguel Silva, P.; Crispim Massuela, D.; Khan, M.W.; Hamar, A.; Khajehei, F.; Graeff-Hönninger, S.; Piatti, C. Quinoa (Chenopodium quinoa Willd.): An Overview of the Potentials of the “Golden Grain” and Socio-Economic and Environmental Aspects of Its Cultivation and Marketization. Foods 2020, 9, 216. [Google Scholar] [CrossRef] [PubMed]
- Bazile, D.; Fuentes, F.; Mujica, A. Historical perspectives and domestication. In Quinoa: Botany, Production and Uses; Bhargava, A., Srivastava, S., Eds.; CABI: Wallingford, UK, 2013; pp. 16–35. [Google Scholar]
- Christensen, S.; Pratt, D.B.; Pratt, C.; Nelson, P.T.; Stevens, M.R.; Jellen, E.N.; Coleman, C.E.; Fairbanks, D.J.; Bonifacio, A.; Maughan, P.J. Assessment of genetic diversity in The USDA and CIP–FAO international nursery collections of quinoa (Chenopodium quinoa Willd.) using microsatellite markers. Plant Genet. Resour. 2007, 5, 82–95. [Google Scholar] [CrossRef]
- Li, G.; Quiros, C.F. Sequence-Related amplified polymorphism (SRAP), a new marker system based on a simple PCR reaction: Its application to mapping and gene tagging in Brassica. Theor. Appl. Genet. 2001, 103, 455–461. [Google Scholar] [CrossRef]
- Salazar, J.; Torres, M.D.L.; Gutierrez, B.; Torres, A.F. Molecular characterization of Ecuadorian quinoa (Chenopodium quinoa Willd.) diversity: Implications for conservation and breeding. Euphytica 2019, 215, 60. [Google Scholar] [CrossRef]
- Zhang, T.; Gu, M.; Liu, Y.; Lv, Y.; Zhou, L.; Lu, H.; Liang, S.; Bao, H.; Zhao, H. Development of novel InDel markers and genetic diversity in Chenopodium quinoa through whole–genome re-sequencing. BMC Genom. 2017, 18, 685. [Google Scholar] [CrossRef] [PubMed]
- Mason, S.; Stevens, M.R.; Jellen, E.N.; Bonifacio, A.; Fairbanks, D.J.; Coleman, C.E.; McCarty, R.R.; Rasmussen, A.G.; Maughan, P.J. Development and use of microsatellite markers for germplasm characterization in quinoa (Chenopodium quinoa Willd.). Crop Sci. 2005, 45, 1618–1630. [Google Scholar] [CrossRef]
- Konishi, Y.; Hirano, S.; Tsuboi, H.; Wada, M. Distribution of minerals in quinoa (Chenopodium quinoa Willd) seeds. Biosci. Biotechnol. Biochem. 2004, 68, 231–234. [Google Scholar] [CrossRef] [PubMed]
- Prado, F.E.; Fernández-Turiel, J.L.; Tsarouchi, M.; Psaras, G.K.; González, J.A. Variation of seed mineral concentrations in seven quinoa cultivars grown in two agroecological sites. Cereal Chem. J. 2014, 91, 453–459. [Google Scholar] [CrossRef]
- Jaime-Pérez, N.; Pineda, B.; García-Sogo, B.; Atares, A.; Athman, A.; Byrt, C.S.; Olías, R.; Asins, M.J.; Gilliham, M.; Moreno, V.; et al. The sodium transporter encoded by the HKT1;2 gene modulates sodium/potassium homeostasis in tomato shoots under salinity. Plant Cell Environ. 2017, 40, 658–671. [Google Scholar] [CrossRef] [PubMed]
- Cai, Z.Q.; Gao, Q. Comparative physiological and biochemical mechanisms of salt tolerance in five contrasting highland quinoa cultivars. BMC Plant Biol. 2020, 20, 70. [Google Scholar] [CrossRef] [PubMed]
- Kiani-Pouya, A.; Rasouli, F.; Shabala, L.; Tahir, A.T.; Zhou, M.; Shabala, S. Understanding the role of root-related traits in salinity tolerance of quinoa accessions with contrasting epidermal bladder cell patterning. Planta 2020, 251, 103. [Google Scholar] [CrossRef]
- Caperta, A.D.; Róis, A.S.; Teixeira, G.; Garcia-Caparros, P.; Flowers, T.J. Secretory structures in plants: Lessons from the Plumbaginaceae on their origin, evolution and roles in stress tolerance. Plant Cell Environ. 2020, 43, 2912–2931. [Google Scholar] [CrossRef]
- Kotula, L.; Garcia Caparros, P.; Zörb, C.; Colmer, T.D.; Flowers, T.J. Improving crop salt tolerance using transgenic approaches: An update and physiological analysis. Plant Cell Environ. 2020, 4, 2932–2956. [Google Scholar] [CrossRef] [PubMed]
- Shabala, S.; Chen, G.; Chen, Z.H.; Pottosin, I. The energy cost of the tonoplast futile sodium leak. New Phytol. 2020, 225, 1105–1110. [Google Scholar] [CrossRef] [PubMed]
- Buhulikar, R.A.; Stanculescu, D.; Preston, C.A.; Baldwin, I.T. ISSR and AFLP analyses of the temporal and spatial population structure of the post-fire annual Nicotiana attenuate in SW, Utah. BMC Ecol. 2004, 4, 12. [Google Scholar]
- Collard, B.C.Y.; Mackill, D.J. Start Codon Targeted (SCoT) Polymorphism: A Simple, Novel DNA Marker Technique for Generating Gene-Targeted Markers in Plants. Plant Mol. Biol. Rep. 2009, 27, 86–93. [Google Scholar] [CrossRef]
- Xiong, F.Q.; Tang, R.H.; Chen, Z.L.; Pan, L.H.; Zhuang, W.J. SCoT: A novel gene targeted marker technique based on the translation start codon. Mol. Plant Breed. 2009, 7, 635–638. [Google Scholar]
- Romero, G.; Adeva, C.; Battad, Z. Genetic fingerprinting: Advancing the frontiers of crop biologyresearch. Philipp. Sci. Lett. 2009, 2, 8–13. [Google Scholar]
- Bafeel, S.O.; Ibrahim, A.A.; Ali, A.A.; Haseeb, A.K.; Anis, A.; Mohammad, A.B. Assessment of DNA barcoding for the identification of Chenopodium murale L. (Chenopodiaceae). Int. J. Biol. Citeseer 2012, 4, 66. [Google Scholar] [CrossRef]
- Ibrahim, S.D.; Abd El-Hakim, A.F.; Ali, H.E.; Abd El-Maksoud, R.M. Genetic differentiation using ISSR, SCoT and DNA Barcoding for Quinoa genotypes. Arab. J. Biotech. 2019, 22, 103–118. [Google Scholar]
- Lema-Rumińska, J.; Miler, N.; Gęsiński, K. Identification of new polish lines of Chenopodium quinoa (Willd.) by spectral analysis of pigments and a confirmation of genetic stability with SCoT and RAPD markers. Acta Sci. Pol. Hortorum Cultus 2018, 17, 75–86. [Google Scholar] [CrossRef]
- Omar, S.A.; Masoud, I.M.; Khalil, R.M.A. Genetic evaluation of some quinoa genotypes under ras suder conditions. J. Plant Prod. Mansoura Univ. 2014, 5, 1915–1930. [Google Scholar]
- Devi, R.J.; Chrungoo, N.K. Species relationships in Chenopodium quinoa and Chenopodium album on the basis of morphology and SDS-PAGE profiles of soluble seed proteins. J. App. Biol. Biotech. 2015, 3, 29–33. [Google Scholar] [CrossRef]
- Ruiz, K.B.; Aloisi, I.; Del Duca, S.; Canelo, V.; Torrigiani, P.; Silva, H.; Biondi, S. Salares versus coastal ecotypes of quinoa: Salinity responses in Chilean landraces from contrasting habitats. Plant Physiol. Biochem. 2016, 101, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Barlett, M.S. Some samples of statistical method of research in agriculture and applied biology. J. R. Stat. Soc. 1973, 4, 137–183. [Google Scholar]
- Radford, P.J. Growth analysis formulae, their use and abuse. Crop Sci. 1967, 7, 171–175. [Google Scholar] [CrossRef]
- Beadle, C.L. Growth analysis. In Photosynthesis and Production in a Changing Environment: A Field and Laboratory Manual; Hall, D.O., Scurlock, J.M.O., Bolhàr-Nordenkampf, H.R., Leegood, R.C., Long, S.P., Eds.; Chapman and Hall: London, UK, 1993; pp. 36–46. [Google Scholar]
- Association of Official Analytical Chemists (AOAC). Official Methods of Analysis of the Association of Official Analytical Chemists, 17th ed.; Association of Official Analytical Chemists: Washington, DC, USA, 2000. [Google Scholar]
- Bates, L.S.; Waldren, R.P.; Teare, I.D. Rapid determination of free proline for water-stress studies. Plant Soil 1973, 39, 205–207. [Google Scholar] [CrossRef]
- Jackson, M.L. Soil Chemical Analysis, 1st ed.; Prentice Hall of India Private Limited: New Delhi, India, 1973. [Google Scholar]
- Cota-Sanchez, H.; Remarchuk, K.; Ubayasenaj, K. Ready-to-use DNA extracted with a CTAB method adapted for herbarium specimens and mucilaginous plant tissue. Plant Mol. Biol. Rep. 2006, 24, 161–167. [Google Scholar] [CrossRef]
- Zietkiewicz, E.; Rafalski, A.; Labuda, D. Genome fingerprinting by simple sequence repeat (SSR)-anchored polymerase chain reaction amplification. Genomics 1994, 20, 176–183. [Google Scholar] [CrossRef] [PubMed]
- Anderson, J.A.; Churchill, G.A.; Autrique, J.E. Optimizing pa-rental selection for genetic linkage maps. Genome 1993, 36, 181–186. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive m method for the quantification of micrograms of protein utilizing the principal dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Maughan, P.J.; Turner, T.B.; Coleman, C.E.; Elzinga, D.B.; Jellen, E.N.; Morales, J.A.; Udall, J.A.; Fairbanks, D.J.; Bonifacio, A. Characterization of salt overly sensitive 1 (SOS1) gene homoeologs in quinoa (Chenopodium quinoa Willd.). Genome 2009, 52, 647–657. [Google Scholar] [CrossRef] [PubMed]
- Morales, A.J.; Bajgain, P.; Garver, Z.; Maughan, P.J.; Udall, J.A. Physiological responses of Chenopodium quinoa to salt stress. Int. J. Plant. Physiol. Biochem. 2011, 3, 219–232. [Google Scholar]
- Balzotti, M.R.B.; Jellen, E.N.; Fairbanks, D.J.; Coleman, C.E.; Stevens, M.R.; Thornton, J.N.; Maughan, P.J.; Mcclellan, D.A. Expression and evolutionary relationships of the Chenopodium quinoa 11S seed storage protein gene. Int. J. Plant Sci. 2008, 169, 281–291. [Google Scholar] [CrossRef]
- Duncan, D.B. Multiple range and multiple F tests. Biometrics 1955, 11, 1–41. [Google Scholar] [CrossRef]
- Hammer, D.A.T.; Ryan, P.D.; Hammer, Ø.; Harper, D.A.T. Past: Paleontological Statistics Software Package for Education and Data Analysis. Palaeontol. Electron. 2001, 4, 1–9. [Google Scholar]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Sokal, R.R.; Michener, C.D. A statistical method for evaluating systematic relationships. Univ. Kans. Sci. Bull. 1958, 38, 1409–1438. [Google Scholar]
- Fuentes, F.F.; Martinez, E.A.; Hinrichsen, P.V.; Jellen, E.N.; Maughan, P.J. Assessment of genetic diversity patterns in Chilean quinoa (Chenopodium quinoa Willd.) germplasm using multiplex fluorescent microsatellite markers. Conserv. Genet. 2009, 10, 369–377. [Google Scholar] [CrossRef]
- Lu, M.; Mo, X.; Wang, Q.; Lu, G.; Jiang, Y. Comparison of Genomic DNA Extraction Methods for Chenopodium quinoa Willd. Agric. Sci. Technol. 2015, 6, 1343. [Google Scholar]
- Rodríguez, A.L.; Isla, M. Comparative analysis of genetic and morphologic diversity among quinoa accessions (Chenopodium quinoa Willd.) of the South of Chile and highland accessions. J. Plant Breed. Crop Sci. 2009, 1, 210–216. [Google Scholar]
- Al-Naggar, A.M.M.; Sobieh, S.E.S.; Atta, M.M.M.; Al-Azab, K.F. Unique SSR markers for drought tolerance in bread wheat mutants derived via exposure to gamma rays. World Res. J. Agron. 2013, 2, 15–25. [Google Scholar]
- Al-Naggar, A.M.M.; Al-Azab, K.F.; Sobieh, S.E.S.; Atta, M.M.M. Molecular analysis of new drought tolerant segregants selected from F2 populations of bread wheat crosses. World Res. J. Agron. 2013, 3, 58–69. [Google Scholar]
- Ana-Cruz, M.C.; Helena, M.E.; Yacenia, M.C. Molecular characterization of Chenopodium quinoa Willd. using inter-simple sequence repeat (ISSR) markers. Afr. J. Biotechnol. 2017, 16, 483–489. [Google Scholar]
- Al-Naggar, M.M.; Abd El-Salam, R.M.; Badran, A.E.E.; Mai El-Moghazi, M.M.A. Molecular Differentiation of Five Quinoa (Chenopodium quinoa Willd.) Genotypes Using Inter-simple Sequence Repeat (ISSR) Markers. Biotechnol. J. Int. 2017, 20, 1–12. [Google Scholar] [CrossRef]
- Saad-Allah, K.M.; Youssef, M.S. Phytochemical and genetic characterization of five quinoa (Chenopodium quinoa Willd.) genotypes introduced to Egypt. Physiol. Mol. Biol. Plants 2018, 24, 617–629. [Google Scholar] [CrossRef] [PubMed]
- Botstein, D.; White, R.L.; Skolnick, M.; Davis, R.W. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 1980, 32, 314–331. [Google Scholar] [PubMed]
- Laosatit, K.; Taytragool, S.; Pimsaythong, K.; Somta, P.; Tanadul, O. Genetic diversity of Quinoa (Chenopodium quinoa Willd.) germplasm as revealed by sequence-related amplified polymorphism markers. Agr. Nat. Resour. 2021, 55, 341–348. [Google Scholar]
- EL-Harty, E.H.; Ghazy, A.; Alateeq, T.K.; Al-Faifi, S.A.; Khan, M.A.; Afzal, M.; Alghamdi, S.S.; Migdadi, H.M. Morphological and Molecular Characterization of Quinoa Genotypes. Agriculture 2021, 11, 286. [Google Scholar] [CrossRef]
- Abd El-Moneim, D. Characterization of ISSR and SCoT Markers and TaWRKY Gene Expression in some Egyptian Wheat Genotypes under Drought Stress. J. Plant Prod. Sci. 2019, 8, 31–46. [Google Scholar]
- Gowayed, S.M.H.; Abd El-Moneim, D. Detection of genetic divergence among some wheat (Triticum aestivum L.) genotypes using molecular and biochemical indicators under salinity stress. PLoS ONE 2021, 16, e0248890. [Google Scholar] [CrossRef] [PubMed]
- EL-Mansy, A.B.; Abd El-Moneim, D.; ALshamrani, S.M.; Alsafhi, F.A.; Abdein, M.A.; Ibrahim, A.A. Genetic Diversity Analysis of Tomato (Solanum lycopersicum L.) with Morphological, Cytological, and Molecular Markers under Heat Stress. Horticulturae 2021, 7, 65. [Google Scholar] [CrossRef]
- Abdein, M.A.; Abd El-Moneim, D.; Taha, S.S.; Al-Juhani, W.S.M.; Mohamed, S.E. Molecular characterization and genetic relationships among some tomato genotypes as revealed by ISSR and SCoT markers. Egypt. J. Genet. Cytol. 2018, 47, 139–159. [Google Scholar]
- Abdein, M.A.; El-Mansy, A.B.; Awad, N.S.; Abd El-Moneim, D. Assessment of Genetic Diversity in Summer Squash Genotypes Using Fruit Morphological, Yield Traits and DNA Markers Analysis under Sinai Conditions. J. Plant Prod. Sci. 2021, 10, 13–29. [Google Scholar]
- Stuardo, M.; Martín, R.S. Antifungal properties of quinoa (Chenopodium quinoa Willd.) alkali treated saponins against botrytis cinerea. Ind. Crop. Prod. 2008, 27, 296–302. [Google Scholar] [CrossRef]
- Sadia, M.; Malik, S.A.; Rabbani, M.A.; Pearce, S.R. Electrophoretic characterization and the relationship between some Brassica species. Electr. J. Biol. 2009, 5, 1–4. [Google Scholar]
- Soliman, M.I.; Ibrahim, A.A.; Rizk, R.M.; Naser, N.S. Phytoremediation, Biochemical and Molecular Studies of Some Selected Hydrophytes in Egypt. J. Appl. Sci. 2019, 19, 708–717. [Google Scholar] [CrossRef]
- Soliman, M.I.; Ibrahim, A.A.; Samaan, L.Z.; Sedky, E. Comparative Studies Between Annual and Perennial Sesbania Using Karyological, Biochemical and Molecular Studies. J. Appl. Sci. 2019, 19, 593–604. [Google Scholar] [CrossRef]
- Aloisi, I.; Parrotta, L.; Ruiz, K.B.; Landi, C.; Bini, L.; Cai, G.; Biondi, S.; Del Duca, S. New insight into quinoa seed quality under salinity: Changes in proteomic and amino acid profiles, phenolic content, and antioxidant activity of protein extracts. Front. Plant Sci. 2016, 7, 656. [Google Scholar] [CrossRef]
- Schmöckel, S.M.; Lightfoot, D.J.; Razali, R.; Tester, M.; Jarvis, D.E. Identification of putative transmembrane proteins involved in salinity tolerance in Chenopodium quinoa by integrating physiological data, RNAseq, and SNP analyses. Front. Plant Sci. 2017, 8, 1023. [Google Scholar] [CrossRef]
- Debez, A.; Hamed, K.B.; Grignon, C.; Abdelly, C. Salinity effects on germination, growth, and seed production of the halophyte Cakile maritima. Plant Soil 2004, 262, 179–189. [Google Scholar] [CrossRef]
- Ruiz-Carrasco, K.; Antognoni, F.; Coulibaly, A.K.; Lizardi, S.; Covarrubias, A.; Martínez, E.A.; Molina-Montenegro, M.A.; Biondi, S.; Zurita-Silva, A. Variation in salinity tolerance of four lowland genotypes of quinoa (Chenopodium quinoa Willd.) as assessed by growth, physiological traits, and sodium transporter gene expression. Plant Physiol. Biochem. 2011, 49, 1333–1341. [Google Scholar] [CrossRef] [PubMed]
- Razzaghi, F.; Jacobsen, S.E.; Jensen, C.R.; Andersen, M.N. Ionic and photosynthetic homeostasis in quinoa challenged by salinity and drought—Mechanisms of tolerance. Func. Plant Biol. 2015, 42, 136–148. [Google Scholar] [CrossRef] [PubMed]
- Tan, M.; Akçay, E. Temperature and salinity effects on germination of some quinoa (Chenopodium quinoa Willd) cultivars. In Proceedings of the 89th IRES International Conference, Helsinki, Finland, 17–18 November 2017; pp. 3–5. [Google Scholar]
- Saleem, M.A.; Basra, S.M.A.; Afzal, I.; Rehman, H.U.; Iqbal, S.; Saddiq, M.S.; Naz, S. Exploring the potential of quinoa accessions for salt tolerance in soilless culture. Int. J. Agric. Biol. 2017, 19, 233–240. [Google Scholar] [CrossRef]
- Long, N. Genetic Variation in Response to Salt Stress of Quinoa Grown under Controlled and Field Conditions. Int. J. Adv. Sci. Eng. Inf. Technol. 2016, 6, 233. [Google Scholar] [CrossRef]
- Talebnejad, R.; Sepaskhah, A.R. Physiological characteristics, gas exchange, and plant ion relations of Quinoa to different saline groundwater depths and water salinity. Arch. Agron. Soil Sci. 2016, 62, 1347–1367. [Google Scholar] [CrossRef]
- Islam, M.T.; Saiful Islam, A.F.M.; Sharaf Uddin, M. Physiological Growth Indices of Maize (Zea mays L.) Genotypes in Sylhet. bioRxiv 2019. [Google Scholar] [CrossRef]
- Riccardi, M.; Pulvento, C.; Lavini, A.; D’Andria, R.; Jacobsen, S.-E. Growth and Ionic Content of Quinoa Under Saline Irrigation. J. Agron. Crop. Sci. 2014, 200, 246–260. [Google Scholar] [CrossRef]
- Jacobsen, S.E.; Quispe, H.; Mujica, A. Quinoa: An alternative crop for saline soils in the Andes. In Scientists and Farmer—Partners in Research for the 21st Century, 2001 CIP Program Report 1999–2000; International Potato Center (Centro Internacional de la Papa) (CIP): Lima, Peru, 2001; pp. 403–408. [Google Scholar]
- Arshadullah, M.; Suhaib, M.; Usama, M.; uz-Zaman, B.; Mahmood, I.A.; Hyder, S.I. Effect of salinity on growth of Chenopodium quinoa Willd. Int. J. Res. Agric. For. 2016, 11, 21–24. [Google Scholar]
- Hussain, M.I.; Muscolo, A.; Ahmed, M.; Asghar, M.A.; Al-Dakheel, A.J. Agro-Morphological, Yield and Quality Traits and Interrelationship with Yield Stability in Quinoa (Chenopodium quinoa Willd.) Genotypes under Saline Marginal Environment. Plants 2020, 9, 1763. [Google Scholar] [CrossRef] [PubMed]
- Miranda, M.; Vega-Gálvez, A.; Martínez, E.A.; López, J.; Marín, J.; Aranda, M.; Fuentes, F. Influence of contrasting environments on seed composition of two quinoa genotypes: Nutritional and functional properties. Chilean J. Agric. Res. 2013, 73, 108–116. [Google Scholar] [CrossRef]
- Hirich, A.; Choukr-Allah, R.; Jacobsen, S.E. Deficit irrigation and organic compost improve growth and yield of Quinoa and pea. J. Agron. Crop. Sci. 2014, 200, 390–398. [Google Scholar] [CrossRef]
- Algosaibi, A.M.; El-Garawany, M.M.; Badran, A.E.; Almadini, A.M. Effect of irrigation water salinity on the growth of quinoa plant. J Agric. Sci. 2015, 7, 205. [Google Scholar] [CrossRef]
- Koyro, H.W.; Eisa, S.S. Effect of salinity on composition, viability, and germination of seeds of Chenopodium quinoa Willd. Plant Soil 2008, 302, 79–90. [Google Scholar] [CrossRef]
- Orsini, F.; Accorsi, M.; Gianquinto, G.; Dinelli, G.; Antognoni, F.; Carrasco, K.B.R.; Martinez, E.A.; Alnayef, M.; Marotti, I.; Bosi, S.; et al. Beyond the ionic and osmotic response to salinity in Chenopodium quinoa: Functional elements of successful halophytism. Funct. Plant Biol. 2011, 38, 818–831. [Google Scholar] [CrossRef] [PubMed]
- Cuin, T.A.; Tian, Y.; Betts, S.A.; Chalmandrier, R.; Shabala, S. Ionic relations and osmotic adjustment in durum and bread wheat under saline conditions. Funct. Plant Biol. 2009, 36, 1110–1119. [Google Scholar] [CrossRef] [PubMed]
- Mansour, M.M.F.; Ali, E.F. Evaluation of proline functions in saline conditions. Phytochemistry 2017, 140, 52–68. [Google Scholar] [CrossRef] [PubMed]
- Prado, F.; Hilal, M.B.; Albornoz, P.L.; Gallardo, M.G.; Ruiz, V.E. Anatomical and physiological responses of four quinoa cultivars to salinity at seedling stage. Indian J. Sci. Technol. 2017, 10, 1–12. [Google Scholar] [CrossRef]
- Derbali, W.; Goussi, R.; Koyro, H.-W.; Abdelly, C.; Manaa, A. Physiological and biochemical markers for screening salt tolerant quinoa genotypes at early seedling stage. J. Plant Interact. 2020, 15, 27–38. [Google Scholar] [CrossRef]
- Shi, H.; Quintero, F.J.; Pardo, J.M.; Zhu, J.-K. The Putative Plasma Membrane Na+/H+ Antiporter SOS1 Controls Long-Distance Na+ Transport in Plants. Plant Cell 2002, 14, 465–477. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Hong, P.; Luo, L.; Xia, T. Overexpression of AtNHX1, a vacuolar Na/H antiporter from Arabidopsis thaliana, in Petunia hybrida enhances salt and drought tolerance. J. Plant Biol. 2009, 52, 453–461. [Google Scholar] [CrossRef]
- Rowley, E.R.; Mockler, T.C. Role of plant transcription factors in biotic stress tolerance. In Abiotic Stress Response in Plants Physiological, Biochemical and Genetic Perspectives; Shanker, A.K., Venkateswarlu, B., Eds.; InTech: Rijeka, Croatia, 2011; pp. 221–268. [Google Scholar]
- Haddoudi, L.; Hdira, S.; Hanana, M.; Romero, I.; Haddoudi, I.; Mahjoub, A.; Ben Jouira, H.; Djébali, N.; Ludidi, N.; Sanchez-Ballesta, M.T.; et al. Evaluation of the Morpho-Physiological, Biochemical and Molecular Responses of Contrasting Medicago truncatula Lines under Water Deficit Stress. Plants 2021, 10, 2114. [Google Scholar] [CrossRef]
- Abd El-Moneim, D.; Alqahtani, M.M.; Abdein, M.A.; Germoush, M.O. Drought and salinity stress response in wheat: Physiological and TaNAC gene expression analysis in contrasting Egyptian wheat genotypes. J. Plant Biotechnol. 2020, 47, 1–14. [Google Scholar] [CrossRef]
- Shabala, S.; Hariadi, Y.; Jacobsen, S.-E. Genotypic difference in salinity tolerance in quinoa is determined by differential control of xylem Na+ loading and stomatal density. J. Plant Physiol. 2013, 170, 906–914. [Google Scholar] [CrossRef] [PubMed]










| Genotype Name | Source |
|---|---|
| M-28 | Denmark |
| Q-37 | Chile |
| S-10 | Denmark |
| Regeolone-3 | Chile |
| LINE-6 | Denmark |
| Molecular Markers | Amplicon Size (bp) Range (bp) | Total No. of Amplicons | Monomorphic Amplicons | Polymorphic Amplicons | Unique Amplicons | % Polymorphism | PIC | |
|---|---|---|---|---|---|---|---|---|
| ISS | ISSR 1 | 196–870 | 29 | 2 | 27 | 17 | 93.1 | 0.81 |
| ISSR 2 | 182–660 | 18 | 2 | 16 | 6 | 88.89 | 0.76 | |
| ISSR 3 | 215–965 | 11 | 1 | 10 | 4 | 90.91 | 0.76 | |
| ISSR 4 | 185–845 | 19 | 2 | 17 | 4 | 89.47 | 0.77 | |
| ISSR 5 | 200–1180 | 19 | 1 | 18 | 8 | 94.74 | 0.81 | |
| ISSR 6 | 145–750 | 17 | 0 | 17 | 4 | 100 | 0.86 | |
| ISSR 7 | 95–970 | 20 | 5 | 15 | 8 | 75 | 0.6 | |
| ISSR 8 | 185–925 | 15 | 0 | 15 | 4 | 100 | 0.88 | |
| ISSR 9 | 80–1430 | 18 | 3 | 15 | 7 | 83.33 | 0.68 | |
| ISSR 10 | 275–1155 | 10 | 0 | 10 | 2 | 100 | 0.77 | |
| Average | - | 32 | 2.91 | 29.09 | 11.64 | 83.30% | 0.77 | |
| SCoT | SCoT 1 | 215–1030 | 14 | 2 | 12 | 5 | 85.71 | 0.76 |
| SCoT 2 | 154–1464 | 20 | 0 | 20 | 5 | 100 | 0.84 | |
| SCoT 3 | 175–840 | 21 | 0 | 21 | 4 | 100 | 0.83 | |
| SCoT 4 | 300–1240 | 15 | 1 | 14 | 6 | 93.33 | 0.81 | |
| SCoT 5 | 155–588 | 10 | 5 | 5 | 2 | 50 | 0.33 | |
| SCoT 6 | 150–790 | 8 | 3 | 3 | 2 | 37.5 | 0.35 | |
| SCoT 7 | 195–1400 | 26 | 0 | 26 | 5 | 100 | 0.82 | |
| SCoT 8 | 132–1900 | 25 | 0 | 25 | 8 | 100 | 0.82 | |
| SCoT 9 | 190–305 | 9 | 5 | 4 | 0 | 44.44 | 0.35 | |
| SCoT 10 | 225–735 | 8 | 5 | 3 | 2 | 37.5 | 0.32 | |
| Average | - | 15.6 | 2.1 | 13.3 | 3.8 | 74.85% | 0.62 | |
| Features | Molecular Markers | SDS-PAGE Protein | ||
|---|---|---|---|---|
| ISSR | SCoT | |||
| Band size range | 80–1430 bp | 132–1900 bp | 12–200 KDa | |
| Total bands | 176 | 156 | 11 | |
| Polymorphic bands | 160 | 133 | 3 | |
| Unique bands | Positive | 19 (M-28), 5(Q-37), 8 (S-10), 4 (Regeolone-3), 28(LINE-6) | 19 (M-28), 1(Q-37), 2 (S-10), 16 (LINE-6) | 3 (M-28) |
| Negative | 1(M-28), 3(LINE-6) | 3 (M-28), 3 (LINE-6) | - | |
| % Polymorphism | 90.91% | 85.26% | 27.27% | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abd El-Moneim, D.; ELsarag, E.I.S.; Aloufi, S.; El-Azraq, A.M.; ALshamrani, S.M.; Safhi, F.A.A.; Ibrahim, A.A. Quinoa (Chenopodium quinoa Willd.): Genetic Diversity According to ISSR and SCoT Markers, Relative Gene Expression, and Morpho-Physiological Variation under Salinity Stress. Plants 2021, 10, 2802. https://doi.org/10.3390/plants10122802
Abd El-Moneim D, ELsarag EIS, Aloufi S, El-Azraq AM, ALshamrani SM, Safhi FAA, Ibrahim AA. Quinoa (Chenopodium quinoa Willd.): Genetic Diversity According to ISSR and SCoT Markers, Relative Gene Expression, and Morpho-Physiological Variation under Salinity Stress. Plants. 2021; 10(12):2802. https://doi.org/10.3390/plants10122802
Chicago/Turabian StyleAbd El-Moneim, Diaa, Eman I. S. ELsarag, Salman Aloufi, Asmaa M. El-Azraq, Salha Mesfer ALshamrani, Fatmah Ahmed Ahmed Safhi, and Amira A. Ibrahim. 2021. "Quinoa (Chenopodium quinoa Willd.): Genetic Diversity According to ISSR and SCoT Markers, Relative Gene Expression, and Morpho-Physiological Variation under Salinity Stress" Plants 10, no. 12: 2802. https://doi.org/10.3390/plants10122802
APA StyleAbd El-Moneim, D., ELsarag, E. I. S., Aloufi, S., El-Azraq, A. M., ALshamrani, S. M., Safhi, F. A. A., & Ibrahim, A. A. (2021). Quinoa (Chenopodium quinoa Willd.): Genetic Diversity According to ISSR and SCoT Markers, Relative Gene Expression, and Morpho-Physiological Variation under Salinity Stress. Plants, 10(12), 2802. https://doi.org/10.3390/plants10122802