Nuclear Localization of HopA1Pss61 Is Required for Effector-Triggered Immunity
Abstract
:1. Introduction
2. Results
2.1. RPS6 (At5g46470) Recognizes HopA1Pss61 in Arabidopsis Col-0 Accession
2.2. HopA1Pss61 Targets to the Nucleus, Cytoplasm, Plasma Membrane, and Plasmodesmata
2.3. Nuclear Localization of HopA1Pss61 Induces Cell Death Responses
2.4. RPS6 Localizes to Nucleus and Cytoplasm
2.5. Induction of HopA1Pss61 Triggers Defense Responses and Bacterial Growth Suppression in Arabidopsis
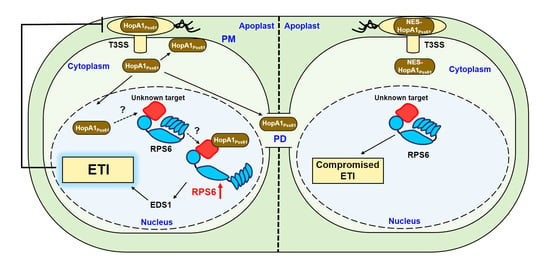
3. Discussion
3.1. Investigation of Genes in RPS6 Locus
3.2. Localization and Functional Analysis of HopA1Pss61 and RPS6
4. Materials and Methods
4.1. Plasmid Construction
4.2. Arabidopsis
4.3. Disease and Bacterial Growth Curve Assay
4.4. HR and Ion Leakage Assays
4.5. Confocal Laser Scanning Microscopy
4.6. Western Blot Analysis and Protein Fractionation
4.7. Yeast Two-Hybrid
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hedrick, S.M. The acquired immune system: A vantage from beneath. Immunity 2004, 21, 607–615. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andolfo, G.; Ercolano, M.R. Plant Innate Immunity Multicomponent Model. Front Plant Sci. 2015, 6, 987. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, J.D.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boller, T.; He, S.Y. Innate immunity in plants: An arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science 2009, 324, 742–744. [Google Scholar] [CrossRef] [Green Version]
- Bigeard, J.; Colcombet, J.; Hirt, H. Signaling mechanisms in pattern-triggered immunity (PTI). Mol Plant 2015, 8, 521–539. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Zhou, J.M. Plant immunity triggered by microbial molecular signatures. Mol Plant 2010, 3, 783–793. [Google Scholar] [CrossRef] [Green Version]
- Chisholm, S.T.; Coaker, G.; Day, B.; Staskawicz, B.J. Host-microbe interactions: Shaping the evolution of the plant immune response. Cell 2006, 124, 803–814. [Google Scholar] [CrossRef] [Green Version]
- Ausubel, F.M. Are innate immune signaling pathways in plants and animals conserved? Nat. Immunol. 2005, 6, 973–979. [Google Scholar] [CrossRef]
- Maekawa, T.; Kufer, T.A.; Schulze-Lefert, P. NLR functions in plant and animal immune systems: So far and yet so close. Nat. Immunol. 2011, 12, 817–826. [Google Scholar] [CrossRef]
- Wang, J.; Hu, M.; Wang, J.; Qi, J.; Han, Z.; Wang, G.; Qi, Y.; Wang, H.W.; Zhou, J.M.; Chai, J. Reconstitution and structure of a plant NLR resistosome conferring immunity. Science 2019, 364. [Google Scholar] [CrossRef]
- Dangl, J.L.; Jones, J.D. Plant pathogens and integrated defence responses to infection. Nature 2001, 411, 826–833. [Google Scholar] [CrossRef]
- Belkhadir, Y.; Subramaniam, R.; Dangl, J.L. Plant disease resistance protein signaling: NBS-LRR proteins and their partners. Curr. Opin. Plant Biol. 2004, 7, 391–399. [Google Scholar] [CrossRef]
- Horsefield, S.; Burdett, H.; Zhang, X.; Manik, M.K.; Shi, Y.; Chen, J.; Qi, T.; Gilley, J.; Lai, J.S.; Rank, M.X.; et al. NAD(+) cleavage activity by animal and plant TIR domains in cell death pathways. Science 2019, 365, 793–799. [Google Scholar] [CrossRef] [Green Version]
- Wan, L.; Essuman, K.; Anderson, R.G.; Sasaki, Y.; Monteiro, F.; Chung, E.H.; Osborne Nishimura, E.; di Antonio, A.; Milbrandt, J.; Dangl, J.L.; et al. TIR domains of plant immune receptors are NAD(+)-cleaving enzymes that promote cell death. Science 2019, 365, 799–803. [Google Scholar] [CrossRef]
- Dodds, P.N.; Lawrence, G.J.; Catanzariti, A.M.; Teh, T.; Wang, C.I.; Ayliffe, M.A.; Kobe, B.; Ellis, J.G. Direct protein interaction underlies gene-for-gene specificity and coevolution of the flax resistance genes and flax rust avirulence genes. Proc. Natl. Acad. Sci. USA 2006, 103, 8888–8893. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.I.; Guncar, G.; Forwood, J.K.; Teh, T.; Catanzariti, A.M.; Lawrence, G.J.; Loughlin, F.E.; Mackay, J.P.; Schirra, H.J.; Anderson, P.A.; et al. Crystal structures of flax rust avirulence proteins AvrL567-A and -D reveal details of the structural basis for flax disease resistance specificity. Plant Cell 2007, 19, 2898–2912. [Google Scholar] [CrossRef] [Green Version]
- Steinbrenner, A.D.; Goritschnig, S.; Staskawicz, B.J. Recognition and activation domains contribute to allele-specific responses of an Arabidopsis NLR receptor to an oomycete effector protein. PLoS Pathog. 2015, 11, e1004665. [Google Scholar] [CrossRef] [Green Version]
- Cesari, S. Multiple strategies for pathogen perception by plant immune receptors. New Phytol. 2018, 219, 17–24. [Google Scholar] [CrossRef]
- Mackey, D.; Holt, B.F., III; Wiig, A.; Dangl, J.L. RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell 2002, 108, 743–754. [Google Scholar] [CrossRef] [Green Version]
- Mackey, D.; Belkhadir, Y.; Alonso, J.M.; Ecker, J.R.; Dangl, J.L. Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell 2003, 112, 379–389. [Google Scholar] [CrossRef] [Green Version]
- Shao, F.; Golstein, C.; Ade, J.; Stoutemyer, M.; Dixon, J.E.; Innes, R.W. Cleavage of Arabidopsis PBS1 by a bacterial type III effector. Science 2003, 301, 1230–1233. [Google Scholar] [CrossRef] [PubMed]
- DeYoung, B.J.; Qi, D.; Kim, S.H.; Burke, T.P.; Innes, R.W. Activation of a plant nucleotide binding-leucine rich repeat disease resistance protein by a modified self protein. Cell Microbiol. 2012, 14, 1071–1084. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.H.; Qi, D.; Ashfield, T.; Helm, M.; Innes, R.W. Using decoys to expand the recognition specificity of a plant disease resistance protein. Science 2016, 351, 684–687. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Dijk, K.; Fouts, D.E.; Rehm, A.H.; Hill, A.R.; Collmer, A.; Alfano, J.R. The Avr (effector) proteins HrmA (HopPsyA) and AvrPto are secreted in culture from Pseudomonas syringae pathovars via the Hrp (type III) protein secretion system in a temperature- and pH-sensitive manner. J. Bacteriol. 1999, 181, 4790–4797. [Google Scholar] [CrossRef] [Green Version]
- Gassmann, W. Natural variation in the Arabidopsis response to the avirulence gene hopPsyA uncouples the hypersensitive response from disease resistance. Mol Plant Microbe Interact. 2005, 18, 1054–1060. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.H.; Kwon, S.I.; Saha, D.; Anyanwu, N.C.; Gassmann, W. Resistance to the Pseudomonas syringae effector HopA1 is governed by the TIR-NBS-LRR protein RPS6 and is enhanced by mutations in SRFR1. Plant Physiol. 2009, 150, 1723–1732. [Google Scholar] [CrossRef] [Green Version]
- Takagi, M.; Hamano, K.; Takagi, H.; Morimoto, T.; Akimitsu, K.; Terauchi, R.; Shirasu, K.; Ichimura, K. Disruption of the MAMP-Induced MEKK1-MKK1/MKK2-MPK4 Pathway Activates the TNL Immune Receptor SMN1/RPS6. Plant Cell Physiol. 2019, 60, 778–787. [Google Scholar] [CrossRef]
- Drechsel, G.; Kahles, A.; Kesarwani, A.K.; Stauffer, E.; Behr, J.; Drewe, P.; Ratsch, G.; Wachter, A. Nonsense-mediated decay of alternative precursor mRNA splicing variants is a major determinant of the Arabidopsis steady state transcriptome. Plant Cell 2013, 25, 3726–3742. [Google Scholar] [CrossRef] [Green Version]
- Gloggnitzer, J.; Akimcheva, S.; Srinivasan, A.; Kusenda, B.; Riehs, N.; Stampfl, H.; Bautor, J.; Dekrout, B.; Jonak, C.; Jimenez-Gomez, J.M.; et al. Nonsense-mediated mRNA decay modulates immune receptor levels to regulate plant antibacterial defense. Cell Host Microbe 2014, 16, 376–390. [Google Scholar] [CrossRef] [Green Version]
- Qi, D.; Dubiella, U.; Kim, S.H.; Sloss, D.I.; Dowen, R.H.; Dixon, J.E.; Innes, R.W. Recognition of the protein kinase AVRPPHB SUSCEPTIBLE1 by the disease resistance protein RESISTANCE TO PSEUDOMONAS SYRINGAE5 is dependent on s-acylation and an exposed loop in AVRPPHB SUSCEPTIBLE1. Plant Physiol. 2014, 164, 340–351. [Google Scholar] [CrossRef] [Green Version]
- Wu, S.W.; Kumar, R.; Iswanto, A.B.B.; Kim, J.Y. Callose balancing at plasmodesmata. J. Exp. Bot. 2018, 69, 5325–5339. [Google Scholar] [CrossRef]
- Iswanto, A.B.; Kim, J.Y. Lipid Raft, Regulator of Plasmodesmal Callose Homeostasis. Plants 2017, 6, 15. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.Y.; Wang, X.; Cui, W.; Sager, R.; Modla, S.; Czymmek, K.; Zybaliov, B.; van Wijk, K.; Zhang, C.; Lu, H.; et al. A plasmodesmata-localized protein mediates crosstalk between cell-to-cell communication and innate immunity in Arabidopsis. Plant Cell 2011, 23, 3353–3373. [Google Scholar] [CrossRef] [Green Version]
- Huang, H.C.; Schuurink, R.; Denny, T.P.; Atkinson, M.M.; Baker, C.J.; Yucel, I.; Hutcheson, S.W.; Collmer, A. Molecular cloning of a Pseudomonas syringae pv. syringae gene cluster that enables Pseudomonas fluorescens to elicit the hypersensitive response in tobacco plants. J. Bacteriol. 1988, 170, 4748–4756. [Google Scholar] [CrossRef] [Green Version]
- Alfano, J.R.; Collmer, A. The type III (Hrp) secretion pathway of plant pathogenic bacteria: Trafficking harpins, Avr proteins, and death. J. Bacteriol. 1997, 179, 5655–5662. [Google Scholar] [CrossRef] [Green Version]
- Garcia, A.V.; Blanvillain-Baufume, S.; Huibers, R.P.; Wiermer, M.; Li, G.; Gobbato, E.; Rietz, S.; Parker, J.E. Balanced nuclear and cytoplasmic activities of EDS1 are required for a complete plant innate immune response. PLoS Pathog. 2010, 6, e1000970. [Google Scholar] [CrossRef] [Green Version]
- Padidam, M. Chemically regulated gene expression in plants. Curr. Opin. Plant Biol. 2003, 6, 169–177. [Google Scholar] [CrossRef]
- Kwon, S.I.; Kim, S.H.; Bhattacharjee, S.; Noh, J.J.; Gassmann, W. SRFR1, a suppressor of effector-triggered immunity, encodes a conserved tetratricopeptide repeat protein with similarity to transcriptional repressors. Plant J. 2009, 57, 109–119. [Google Scholar] [CrossRef]
- Bhattacharjee, S.; Halane, M.K.; Kim, S.H.; Gassmann, W. Pathogen effectors target Arabidopsis EDS1 and alter its interactions with immune regulators. Science 2011, 334, 1405–1408. [Google Scholar] [CrossRef]
- Kim, S.H.; Gao, F.; Bhattacharjee, S.; Adiasor, J.A.; Nam, J.C.; Gassmann, W. The Arabidopsis resistance-like gene SNC1 is activated by mutations in SRFR1 and contributes to resistance to the bacterial effector AvrRps4. PLoS Pathog. 2010, 6, e1001172. [Google Scholar] [CrossRef]
- Saucet, S.B.; Ma, Y.; Sarris, P.F.; Furzer, O.J.; Sohn, K.H.; Jones, J.D. Two linked pairs of Arabidopsis TNL resistance genes independently confer recognition of bacterial effector AvrRps4. Nat. Commun. 2015, 6, 6338. [Google Scholar] [CrossRef]
- Halane, M.K.; Kim, S.H.; Spears, B.J.; Garner, C.M.; Rogan, C.J.; Okafor, E.C.; Su, J.; Bhattacharjee, S.; Gassmann, W. The bacterial type III-secreted protein AvrRps4 is a bipartite effector. PLoS Pathog. 2018, 14, e1006984. [Google Scholar] [CrossRef]
- Sarris, P.F.; Duxbury, Z.; Huh, S.U.; Ma, Y.; Segonzac, C.; Sklenar, J.; Derbyshire, P.; Cevik, V.; Rallapalli, G.; Saucet, S.B.; et al. A Plant Immune Receptor Detects Pathogen Effectors that Target WRKY Transcription Factors. Cell 2015, 161, 1089–1100. [Google Scholar] [CrossRef] [Green Version]
- Cesari, S.; Thilliez, G.; Ribot, C.; Chalvon, V.; Michel, C.; Jauneau, A.; Rivas, S.; Alaux, L.; Kanzaki, H.; Okuyama, Y.; et al. The rice resistance protein pair RGA4/RGA5 recognizes the Magnaporthe oryzae effectors AVR-Pia and AVR1-CO39 by direct binding. Plant Cell 2013, 25, 1463–1481. [Google Scholar] [CrossRef] [Green Version]
- Cesari, S.; Kanzaki, H.; Fujiwara, T.; Bernoux, M.; Chalvon, V.; Kawano, Y.; Shimamoto, K.; Dodds, P.; Terauchi, R.; Kroj, T. The NB-LRR proteins RGA4 and RGA5 interact functionally and physically to confer disease resistance. Embo J. 2014, 33, 1941–1959. [Google Scholar] [CrossRef]
- Takagi, M.; Iwamoto, N.; Kubo, Y.; Morimoto, T.; Takagi, H.; Takahashi, F.; Nishiuchi, T.; Tanaka, K.; Taji, T.; Kaminaka, H.; et al. Arabidopsis SMN2/HEN2, Encoding DEAD-Box RNA Helicase, Governs Proper Expression of the Resistance Gene SMN1/RPS6 and Is Involved in Dwarf, Autoimmune Phenotypes of mekk1 and mpk4 Mutants. Plant Cell Physiol. 2020, 61, 1507–1516. [Google Scholar] [CrossRef]
- Aung, K.; Xin, X.; Mecey, C.; He, S.Y. Subcellular Localization of Pseudomonas syringae pv. tomato Effector Proteins in Plants. Methods Mol Biol. 2017, 1531, 141–153. [Google Scholar] [CrossRef] [Green Version]
- Burch-Smith, T.M.; Schiff, M.; Caplan, J.L.; Tsao, J.; Czymmek, K.; Dinesh-Kumar, S.P. A novel role for the TIR domain in association with pathogen-derived elicitors. PLoS Biol. 2007, 5, e68. [Google Scholar] [CrossRef]
- Wirthmueller, L.; Zhang, Y.; Jones, J.D.; Parker, J.E. Nuclear accumulation of the Arabidopsis immune receptor RPS4 is necessary for triggering EDS1-dependent defense. Curr. Biol. 2007, 17, 2023–2029. [Google Scholar] [CrossRef] [Green Version]
- Carmo-Fonseca, M.; Mendes-Soares, L.; Campos, I. To be or not to be in the nucleolus. Nat. Cell Biol. 2000, 2, E107–E112. [Google Scholar] [CrossRef]
- Fuhrman, L.E.; Goel, A.K.; Smith, J.; Shianna, K.V.; Aballay, A. Nucleolar proteins suppress Caenorhabditis elegans innate immunity by inhibiting p53/CEP-1. PLoS Genet. 2009, 5, e1000657. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, Y.; Huang, D.; Chen, X. Dynamic regulation of plasmodesmatal permeability and its application to horticultural research. Hortic. Res. 2019, 6, 47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luna, E.; Pastor, V.; Robert, J.; Flors, V.; Mauch-Mani, B.; Ton, J. Callose deposition: A multifaceted plant defense response. Mol Plant Microbe Interact. 2011, 24, 183–193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malinovsky, F.G.; Fangel, J.U.; Willats, W.G. The role of the cell wall in plant immunity. Front. Plant Sci. 2014, 5, 178. [Google Scholar] [CrossRef] [Green Version]
- Blumke, A.; Falter, C.; Herrfurth, C.; Sode, B.; Bode, R.; Schafer, W.; Feussner, I.; Voigt, C.A. Secreted fungal effector lipase releases free fatty acids to inhibit innate immunity-related callose formation during wheat head infection. Plant Physiol. 2014, 165, 346–358. [Google Scholar] [CrossRef] [Green Version]
- Bartetzko, V.; Sonnewald, S.; Vogel, F.; Hartner, K.; Stadler, R.; Hammes, U.Z.; Bornke, F. The Xanthomonas campestris pv. vesicatoria type III effector protein XopJ inhibits protein secretion: Evidence for interference with cell wall-associated defense responses. Mol Plant Microbe Interact. 2009, 22, 655–664. [Google Scholar] [CrossRef] [Green Version]
- Sohn, K.H.; Zhang, Y.; Jones, J.D. The Pseudomonas syringae effector protein, AvrRPS4, requires in planta processing and the KRVY domain to function. Plant J. 2009, 57, 1079–1091. [Google Scholar] [CrossRef]
- Zaman, N.; Seitz, K.; Kabir, M.; George-Schreder, L.S.; Shepstone, I.; Liu, Y.; Zhang, S.; Krysan, P.J. A Forster resonance energy transfer sensor for live-cell imaging of mitogen-activated protein kinase activity in Arabidopsis. Plant J. 2019, 97, 970–983. [Google Scholar] [CrossRef]
- Gassmann, W.; Hinsch, M.E.; Staskawicz, B.J. The Arabidopsis RPS4 bacterial-resistance gene is a member of the TIR-NBS-LRR family of disease-resistance genes. Plant J. 1999, 20, 265–277. [Google Scholar] [CrossRef]
- Serrano, I.; Gu, Y.; Qi, D.; Dubiella, U.; Innes, R.W. The Arabidopsis EDR1 protein kinase negatively regulates the ATL1 E3 ubiquitin ligase to suppress cell death. Plant Cell 2014, 26, 4532–4546. [Google Scholar] [CrossRef] [Green Version]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kang, H.; Nguyen, Q.-M.; Iswanto, A.B.B.; Hong, J.C.; Bhattacharjee, S.; Gassmann, W.; Kim, S.H. Nuclear Localization of HopA1Pss61 Is Required for Effector-Triggered Immunity. Plants 2021, 10, 888. https://doi.org/10.3390/plants10050888
Kang H, Nguyen Q-M, Iswanto ABB, Hong JC, Bhattacharjee S, Gassmann W, Kim SH. Nuclear Localization of HopA1Pss61 Is Required for Effector-Triggered Immunity. Plants. 2021; 10(5):888. https://doi.org/10.3390/plants10050888
Chicago/Turabian StyleKang, Hobin, Quang-Minh Nguyen, Arya Bagus Boedi Iswanto, Jong Chan Hong, Saikat Bhattacharjee, Walter Gassmann, and Sang Hee Kim. 2021. "Nuclear Localization of HopA1Pss61 Is Required for Effector-Triggered Immunity" Plants 10, no. 5: 888. https://doi.org/10.3390/plants10050888
APA StyleKang, H., Nguyen, Q. -M., Iswanto, A. B. B., Hong, J. C., Bhattacharjee, S., Gassmann, W., & Kim, S. H. (2021). Nuclear Localization of HopA1Pss61 Is Required for Effector-Triggered Immunity. Plants, 10(5), 888. https://doi.org/10.3390/plants10050888