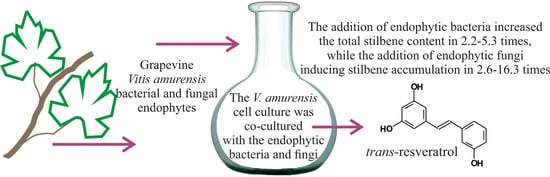
The Influence of the Grapevine Bacterial and Fungal Endophytes on Biomass Accumulation and Stilbene Production by the In Vitro Cultivated Cells of Vitis amurensis Rupr.
Abstract
:1. Introduction
2. Results
2.1. Identification of V. amurensis Endophytes
2.2. The Influence of V. amurensis Endophytes on the Fresh Biomass Accumulation in V. amurensis Cell Culture
2.3. The Effect of the Endophytic Bacteria and Fungi on Stilbene Content in the V. amurensis Cell Suspension
2.4. VaPAL and VaSTS Gene Expression in V. amurensis Cells with the Addition of Endophytes
2.5. Sensitivity of the Grape Endophytes to Antibiotics, Fluconazole, and Resveratrol
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Plant Material
5.2. Isolation and Identification of the Endophytic Bacteria and Fungi
5.3. Treatment of Grape Cells with Endophytic Bacteria and Fungi
5.4. Total RNA Extraction, Reverse Transcription, and qRT-PCR
5.5. High-Performance Liquid Chromatography
5.6. Antibiotic Susceptibility Analysis
5.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Nair, D.N.; Padmavathy, S. Impact of Endophytic Microorganisms on Plants, Environment and Humans. Hindawi Publ. Corpor. Sci. World J. 2014, 2014, 250693. [Google Scholar] [CrossRef] [Green Version]
- Salvetti, E.; Campanaro, S.; Campedelli, I.; Fracchetti, F.; Gobbi1, A.; Tornielli, G.B.; Torriani, S.; Felis, G.E. Whole-metagenome-sequencing based community profiles of Vitis vinifera L. cv. Corvina berries withered in two post-harvest conditions. Front. Microbiol. 2016, 7, 937. [Google Scholar] [CrossRef] [Green Version]
- Jayawardena, R.S.; Purahong, W.; Zhang, W.; Wubet, T.; Li, X.H.; Liu, M.; Zhao, W.; Hyde, K.D.; Liu, J.H.; Yan, J. Biodiversity of fungi on Vitis vinifera L. revealed by traditional and high-resolution culture-independent approaches. Fun Diver. 2018, 90, 1–84. [Google Scholar] [CrossRef] [Green Version]
- Gamalero, E.; Bona, E.; Novello, G.; Boatti, L.; Mignone, F.; Massa, N.; Cesaro, P.; Berta, G.; Lingua, G. Discovering the bacteriome of Vitis vinifera cv. Pinot Noir in a conventionally managed vineyard. Sci. Rep. 2020, 10, 6453. [Google Scholar] [CrossRef] [Green Version]
- Campisano, A.; Pancher, M.; Puopolo, G.; Puddu, A.; Lopez-Fernandez, S.; Biagini, B.; Yousaf, S.; Pertot, I. Diversity in endophyte populations reveals functional and taxonomic diversity between wild and domesticated grapevines. Am. J. Enol. Vitic. 2015, 66, 12–21. [Google Scholar] [CrossRef]
- Andreolli, M.; Lampis, S.; Zapparoli, G.; Angelini, E.; Vallini, G. Diversity of bacterial endophytes in 3- and 15-year-old grapevines of Vitis vinifera cv. Corvina and their potential for plant growth promotion and phytopathogen control. Microb. Res. 2016, 183, 42–52. [Google Scholar] [CrossRef]
- Theocharis, A.; Bordiec, S.; Fernandez, O.; Paquis, S.; Dhondt-Cordelier, S.; Baillieul, F.; Clément, C.; Barka, E.A. Burkholderia phytofirmans PsJN primes Vitis vinifera L. and confers a better tolerance to low nonfreezing temperatures. Mol. Plant Microbe Interact. 2012, 25, 241–249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verhagen, B.; Trotel-Aziz, P.; Jeandet, P.; Baillieul, F.; Aziz, A. Improved resistance against Botrytis cinerea by grapevine-associated bacteria that induce a prime oxidative burst and phytoalexin production. Phytopathology 2011, 101, 768–777. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, M.; Chen, L.C.; Qu, J.Z.; Liu, F.; Zhou, M.; Ma, Y.M.; Xiang, S.Y.; Pan, X.X.; Zhang, H.B.; Yang, M.Z. Exposure to endophytic fungi quantitatively and compositionally alters anthocyanins in grape cells. Plant Physiol. Biochem. 2020, 149, 144–152. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.Z.; Huang, L.H.; Ao, X.J.; Ren, A.Y.; Yuan, M.Q.; Zhang, H.B. Endophytic fungal strains specifically modified the biochemical status of grape cells. J. Plant Biol. 2018, 61, 210–216. [Google Scholar] [CrossRef]
- Chen, Q.; Diao, L.; Song, H.; Zhu, X. Vitis amurensis Rupr: A review of chemistry and pharmacology. Phytomedicine 2018, 49, 111–122. [Google Scholar] [CrossRef]
- Wang, Y.; Xin, H.; Fan, P.; Zhang, J.; Liu, Y.; Dong, Y.; Wang, Z.; Yang, Y.; Zhang, Q.; Ming, R.; et al. The genome of Shanputao (Vitis amurensis) provides a new insight into cold tolerance of grapevine. Plant J. 2021, 105, 1495–1506. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Zhang, Y.; Zhang, H.; Huang, H.; Folta, K.; Lu, J. Whole genome wide expression profiles of Vitis amurensis grape responding to downy mildew by using Solexa sequencing technology. BMC Plant Biol. 2010, 10, 234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, L.; Li, H. Research progress in amur grape, Vitis amurensis Rupr. Can. J. Plant Sci. 2013, 93, 565–575. [Google Scholar] [CrossRef] [Green Version]
- Yim, N.; Ha, T.; Trung, T.; Kim, J.; Lee, S.; Na, M.; Jung, H.; Kim, H.; Kim, Y.; Bae, K. The antimicrobial activity of compounds from the leaf and stem of Vitis amurensis against two oral pathogens. Bioorg. Med. Chem. Lett. 2010, 20, 1165–1168. [Google Scholar] [CrossRef]
- Wang, C.; Ai, J.; Liu, Y.; Lv, H.; Fan, S.; Yang, Y. Fusarium avenaceum: A New Pathogen Causing Amur Grape (Vitis amurensis) Fruit Rot in Jilin Province, China. Plant Dis. 2015, 99, 889. [Google Scholar] [CrossRef]
- Li, Z.; Chang, P.; Gao, L.; Wang, X. The Endophytic Fungus Albifimbria verrucaria from Wild Grape as an Antagonist of Botrytis cinerea and Other Grape Pathogens. Phytopathology 2020, 110, 843–850. [Google Scholar] [CrossRef]
- Chong, J.; Poutaraud, A.; Hugueney, P. Metabolism and roles of stilbenes in plants. Plant Sci. 2009, 177, 143–155. [Google Scholar] [CrossRef]
- Kiselev, K.V. Perspectives for production and application of resveratrol. Appl. Microbiol. Biotechnol. 2011, 90, 417–425. [Google Scholar] [CrossRef] [PubMed]
- Suwalsky, M.; Villena, F.; Gallardo, M.J. In vitro protective effects of resveratrol against oxidative damage in human erythrocytes. Biochim. Biophys. Acta Biomembr. 2015, 1848, 76–82. [Google Scholar] [CrossRef] [Green Version]
- Jeandet, P.; Douillet-Breuil, A.C.; Bessis, R.; Debord, S.; Sbaghi, M.; Adrian, M. Phytoalexins from the Vitaceae: Biosynthesis, Phytoalexin Gene Expression in Transgenic Plants, Antifungal Activity, and Metabolism. J. Agric. Food Chem. 2002, 50, 2731–2741. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, J.H.; Ferraro, M.J. Antimicrobial Susceptibility Testing: A Review of General Principles and Contemporary Practices. Med. Microbiol. 2009, 49, 1749–1755. [Google Scholar] [CrossRef]
- Ma, D.S.L.; Tan, L.T.H.; Chan, K.G.; Yap, W.H.; Pusparajah, P.; Chuah, L.H.; Ming, L.C.; Khan, T.M.; Lee, L.H.; Goh, B.H. Resveratrol—Potential Antibacterial Agent against Foodborne Pathogens. Front. Pharmacol. 2018, 9, 102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, L.H.; Yuan, M.Q.; Ao, X.J.; Ren, A.Y.; Zhang, H.B.; Yang, M.Z. Endophytic fungi specifically introduce novel metabolites into grape flesh cells in vitro. PLoS ONE 2018, 13, e0196996. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramirez-Suero, M.; Bénard-Gellon, M.; Chong, J.; Laloue, H.; Stempien, E.; Abou-Mansour, E.; Fontaine, F.; Larignon, P.; Mazet-Kieffer, F.; Farine, S. Extracellular compounds produced by fungi associated with Botryosphaeria dieback induce differential defence gene expression patterns and necrosis in Vitis vinifera cv. Chardonnay cells. Protoplasma 2014, 251, 1417–1426. [Google Scholar] [CrossRef] [Green Version]
- Dubrovina, A.S.; Kiselev, K.V. Regulation of stilbene biosynthesis in plants. Planta 2017, 346, 597–623. [Google Scholar] [CrossRef]
- Ku, K.L.; Chang, P.S.; Cheng, Y.C.; Lien, C.Y. Production of stilbenoids from the callus of Arachis hypogaea: A novel source of the anticancer compound piceatannol. J. Agric. Food. Chem. 2005, 53, 3877–3881. [Google Scholar] [CrossRef]
- Yang, M.H.; Kuo, C.H.; Hsieh, W.C.; Ku, K.L. Investigation of microbial elicitation of trans-resveratrol and trans-piceatannol in peanut callus led to the application of chitin as a potential elicitor. J. Agric. Food. Chem. 2010, 58, 9537–9541. [Google Scholar] [CrossRef]
- Xu, A.; Zhan, J.C.; Huang, W.D. Effects of ultraviolet C, methyl jasmonate and salicylic acid, alone or in combination, on stilbene biosynthesis in cell suspension cultures of Vitis vinifera L. cv. Cabernet Sauvignon. Plant Cell Tissue Organ Cult. 2015, 122, 197–211. [Google Scholar] [CrossRef]
- Larronde, F.; Gaudillère, J.P.; Krisa, S.; Decendit, A.; Deffieux, G.; Mérillon, J.M. Airborne methyl jasmonate induces stilbene accumulation in leaves and berries of grapevine plants. Am. J. Enol. Viti. 2003, 54, 60–63. [Google Scholar]
- Kiselev, K.V.; Aleynova, O.A.; Grigorchuk, V.P.; Dubrovina, A.S. Stilbene accumulation and expression of stilbene biosynthesis pathway genes in wild grapevine Vitis amurensis Rupr. Planta 2017, 245, 151–159. [Google Scholar] [CrossRef] [PubMed]
- Belchí-Navarro, S.; Almagro, L.; Sabater-Jara, A.B.; Fernández-Pérez, F.; Bru, R.; Pedreño, M.A. Induction of trans-resveratrol and extracellular pathogenesis-related proteins in elicited suspension cultured cells of Vitis vinifera cv Monastrell. J. Plant Physiol. 2013, 170, 258–264. [Google Scholar] [CrossRef] [PubMed]
- Almagro, L.; Belchi-Navarro, S.; Martinez-Marquez, A.; Bru, R.; Pedreno, M.A. Enhanced extracellular production of trans-resveratrol in Vitis vinifera suspension cultured cells by using cyclodextrins and coronatine. Plant Physiol. Biochem. 2015, 97, 361–367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reasoner, D.J.; Geldreich, E.E. A new medium for the enumeration and subculture of bacteria from potable water. Appl. Environ. Microbiol. 1985, 49, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kiselev, K.V.; Ageenko, N.V.; Kurilenko, V.V. Involvement of the cell-specific pigment genes pks and sult in the bacteria defense response of the sea urchin Strongylocentrotus intermedius. Dis. Aquat. Org. 2013, 103, 121–132. [Google Scholar] [CrossRef] [Green Version]
- Lane, D.J. 16S/23S rRNA Sequencing. In Nucleic Acid Techniques in Bacterial Systematic; Stackebrandt, E., Goodfellow, M., Eds.; John Wiley and Sons: New York, NY, USA, 1991; pp. 115–175. [Google Scholar]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Academic Press, Inc.: Cambridge, MA, USA, 1990; pp. 315–322. [Google Scholar]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Aligment Search Tool. J. Mol. Biol. 1990, 3, 403–410. [Google Scholar] [CrossRef]
- Tyunin, A.P.; Suprun, A.R.; Nityagovsky, N.N.; Manyakhin, A.Y.; Karetin, Y.A.; Dubrovina, A.S.; Kiselev, K.V. The effect of explant origin and collection season on stilbene biosynthesis in cell cultures of Vitis amurensis Rupr. Plant Cell Tiss. Organ Cult. 2019, 136, 189–196. [Google Scholar] [CrossRef]
- Dubrovina, A.S.; Kiselev, K.V. Effect of long-term cultivation on resveratrol accumulation in a high-producing cell culture of Vitis amurensis. Acta Physiol. Plant. 2012, 34, 1101–1106. [Google Scholar] [CrossRef]
- Kiselev, K.V.; Aleynova, O.A.; Tyunin, A.P. Expression of the R2R3 MYB transcription factors in Vitis amurensis Rupr. plants and cell cultures with different resveratrol content. Russ. J. Genet. 2017, 53, 465–471. [Google Scholar] [CrossRef]
- Kiselev, K.V.; Dubrovina, A.S.; Shumakova, O.A.; Karetin, Y.A.; Manyakhin, A.Y. Structure and expression profiling of a novel calcium-dependent protein kinase gene, CDPK3a, in leaves, stems, grapes, and cell cultures of wild-growing grapevine Vitis amurensis Rupr. Plant Cell Rep. 2013, 32, 431–442. [Google Scholar] [CrossRef]
- Dubrovina, A.S.; Kiselev, K.V.; Khristenko, V.S.; Aleynova, O.A. VaCPK20, a calcium-dependent protein kinase gene of wild grapevine Vitis amurensis Rupr., mediates cold and drought stress tolerance. J. Plant Physiol. 2015, 185, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Reid, K.E.; Olsson, N.; Schlosser, J.; Peng, F.; Lund, S.T. An optimized grapevine RNA isolation procedure and statistical determination of reference genes for real-time RT-PCR during berry development. BMC Plant Biol. 2006, 6, 27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta (T)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]



| № | Used Gene | Genus and Sequence ID | The Close Species and Sequence ID | Percent Identity |
|---|---|---|---|---|
| 1 | 16S rRNA | Agrobacterium (MZ424738) | Agrobacterium rubi (MN752429.1) | 99.17% |
| 2 | 16S rRNA | Bacillus (MZ424739) | Bacillus thuringiensis (KU179338.1) | 100% |
| 3 | 16S rRNA | Curtobacterium (MZ424740) | Curtobacterium flaccumfaciens (AJ310414.1) | 100% |
| 4 | 16S rRNA | Erwinia (MZ424741) | Erwinia billingiae (KM408608.1) | 100% |
| 5 | 16S rRNA | Pantoae (MZ424742) | Pantoea agglomerans (MT605813.1) | 99.75% |
| 6 | 16S rRNA | Pseudomonas (MZ424743) | Pseudomonas alkylphenolica (MN813762.1) | 99.89% |
| 7 | 16S rRNA | Xanthomonas (MZ424744) | Xanthomonas campestris (MN108237.1) | 99.13% |
| 8 | ITS1 | Alternaria (MZ427922) | Alternaria tenuissima (KF308883.1) | 100% |
| 9 | ITS1 | Biscogniauxia (MZ427923) | Biscogniauxia maritima (MN341558.1) | 100% |
| 10 | ITS1 | Cladosporium (MZ427924) | Cladosporium perangustum (MT645918.1) | 100% |
| 11 | ITS1 | Didymella (MZ427925) | Didymella negriana (MK100201.1) | 100% |
| 12 | ITS1 | Didymella (MZ427926) | Didymella pinodella (KX869956.1) | 100% |
| 13 | ITS1 | Fusarium (MZ427927) | Fusarium tricinctum (MT446111.1) | 100% |
| 14 | ITS1 | Trichoderma (MZ427928) | Trichoderma harzianum (MT422092.1) | 98.97% |
| Di-glucoside trans-Resveratrol | trans-Piceid | trans-Resveratrol | epsilon-Viniferin | delta-Viniferin | cis-Resveratrol | cis-Piceid | trans-Piceatannol | |
|---|---|---|---|---|---|---|---|---|
| V7, Control | 0.432 ± 0.135 | 0.104 ± 0.022 | 0.029 ± 0.008 | 0.024 ± 0.007 | 0.255 ± 0.077 | 0.0004 ± 0.0001 | 0.0103 ± 0.0061 | 0 |
| Agrobacterium sp. | 0.583 ± 0.167 | 0.136 ± 0.055 | 0.140 ** ± 0.036 | 0.279 * ± 0.129 | 0.802 * ± 0.240 | 0.0010 ± 0.0004 | 0.0017 ± 0.0015 | 0 |
| Bacillus sp. | 0.823 ± 0.322 | 0.291 ** ± 0.053 | 0.141 ** ± 0.049 | 0.190 ** ± 0.068 | 0.575 ± 0.213 | 0.0011 *± 0.0004 | 0.0693 ± 0.0561 | 0.0014 ± 0.0014 |
| Curtobacterium sp. | 3.447 * ± 0.247 | 0.348 ** ± 0.082 | 0.181 ** ± 0.043 | 0.131 ** ± 0.024 | 1.412 ** ± 0.415 | 0.0015 ** ± 0.0003 | 0.1025 ** ± 0.0674 | 0 |
| Erwinia sp. | 0.860 ± 0.350 | 0.243 ± 0.078 | 0.364 * ± 0.201 | 1.154 ** ± 0.535 | 0.940 * ± 0.333 | 0.0018 * ± 0.0008 | 0.0143 ± 0.0059 | 0.0249 * ± 0.0228 |
| Pantoae sp. | 0.602 ± 0.122 | 0.194 ± 0.074 | 0.477 ** ± 0.291 | 0.744 ** ± 0.204 | 0.770 ± 0.292 | 0.0010 ± 0.0003 | 0.0117 ± 0.0050 | 0.0013 ± 0.0013 |
| Pseudomonas sp. | 0.486 ± 0.232 | 0.230 ± 0.094 | 0.167 ** ± 0.047 | 0.440 ** ± 0.158 | 0.861 * ± 0.304 | 0.0016 ± 0.0007 | 0.0084 ± 0.0045 | 0.0245 ± 0.0245 |
| Xantomonas sp. | 4.491 * ±2.124 | 0.364 ** ± 0.148 | 0.433 ** ± 0.012 | 0.372 ** ± 0.220 | 1.958 * ± 1.605 | 0.0022 ** ± 0.0001 | 0.0268 ± 0.0268 | 0 |
| Alternaria sp. | 0.620 ± 0.255 | 0.239 ± 0.098 | 0.338 ** ± 0.117 | 1.292 ** ± 0.586 | 0.941 ± 0.469 | 0.0029 ± 0.0013 | 0.0501 ± 0.0326 | 0.0046 ± 0.0046 |
| Biscogniauxia sp. | 1.265 * ± 0.206 | 0.477 ** ± 0.106 | 2.861 ** ± 0.415 | 3.952 ** ± 0.664 | 4.981 ** ± 0.335 | 0.0035 * ± 0.0002 | 0.0910 ± 0.0162 | 0 |
| Cladosporium sp. | 1.258 ** ± 0.178 | 0.346 ** ± 0.082 | 0.882 ** ± 0.136 | 2.964 ** ± 0.132 | 1.504 * ± 0.866 | 0.0040 ** ± 0.0002 | 0.0216 ± 0.0125 | 0 |
| Didymella sp. 1 | 1.035 ± 0.667 | 0.427 * ± 0.339 | 1.180 * ± 1.130 | 2.082 ** ± 1.872 | 2.222 * ± 2.060 | 0.002 ± 0.002 | 0 | 0 |
| Didymella sp. 2 | 2.184 ** ± 0.945 | 0.619 ** ± 0.077 | 2.796 ** ± 1.856 | 3.860 ** ± 0.076 | 3.994 ** ± 0.775 | 0.005 ** ± 0.001 | 0.298 ** ± 0.169 | 0 |
| Fusarium sp. | 0.487 ± 0.236 | 0.147 ± 0.078 | 0.171 * ± 0.073 | 0.614 ** ± 0.163 | 0.700 * ± 0.229 | 0.0173 ** ± 0.0165 | 0.0295 ± 0.0184 | 0 |
| Trichoderma sp. | 0.682 ± 0.024 | 0.099 ± 0.001 | 0.127 ** ± 0.015 | 0.195 ** ± 0.079 | 2.706 ** ± 0.884 | 0.0025 ** ± 0.0004 | 0.0465 ± 0.0268 | 0.0094 * ± 0.0054 |
| Antibiotics | Res. | Fluc. | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Genus | Ap | Cam | Cf | Gent | Km | Rf | Sp | Tet | ||
| Agrobacterium | 0.3 | 0 | 1 | 1 | 1.2 | 0.5 | 1 | 1.6 | 0 | n.m. |
| Bacillus | 0 | 0 | 0 | 0.6 | 0.5 | 0.6 | 0 | 0 | 0 | n.m. |
| Erwinia | 0 | 0 | 1 | 0.5 | 0.6 | 0.3 | 0 | 0 | 0.2 | n.m. |
| Pantoae | 0 | 0 | 0 | 0.6 | 0.8 | 0 | 0 | 0 | 0 | n.m. |
| Curtobacterium | 0 | 0 | 0 | 0.8 | 1 | 2 | 0 | 0.2 | 0 | n.m. |
| Xantomonas | 0 | 0 | 0 | 0.6 | 0.8 | 0.8 | 0 | 0 | 0 | n.m. |
| Pseudomonas | 0 | 0 | 0 | 0.6 | 0.5 | 0.7 | 0.6 | 0.7 | 0 | n.m. |
| Fusarium | n.m. | n.m. | n.m. | n.m. | n.m. | n.m. | n.m. | n.m. | 0 * | 0.1 * |
| Alternaria | n.m. | n.m. | n.m. | n.m. | n.m. | n.m. | n.m. | n.m. | 0 * | 0.1 * |
| Didymella-1 | n.m. | n.m. | n.m. | n.m. | n.m. | n.m. | n.m. | n.m. | 0 * | 0 * |
| Didymella-2 | n.m. | n.m. | n.m. | n.m. | n.m. | n.m. | n.m. | n.m. | 0 * | 0 * |
| Trichoderma | n.m. | n.m. | n.m. | n.m. | n.m. | n.m. | n.m. | n.m. | 0 | 0 * |
| Cladosporium | n.m. | n.m. | n.m. | n.m. | n.m. | n.m. | n.m. | n.m. | 0 * | 0.5 |
| Biscogniauxia | n.m. | n.m. | n.m. | n.m. | n.m. | n.m. | n.m. | n.m. | 0 * | 0 * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aleynova, O.A.; Suprun, A.R.; Nityagovsky, N.N.; Dubrovina, A.S.; Kiselev, K.V. The Influence of the Grapevine Bacterial and Fungal Endophytes on Biomass Accumulation and Stilbene Production by the In Vitro Cultivated Cells of Vitis amurensis Rupr. Plants 2021, 10, 1276. https://doi.org/10.3390/plants10071276
Aleynova OA, Suprun AR, Nityagovsky NN, Dubrovina AS, Kiselev KV. The Influence of the Grapevine Bacterial and Fungal Endophytes on Biomass Accumulation and Stilbene Production by the In Vitro Cultivated Cells of Vitis amurensis Rupr. Plants. 2021; 10(7):1276. https://doi.org/10.3390/plants10071276
Chicago/Turabian StyleAleynova, Olga A., Andrey R. Suprun, Nikolay N. Nityagovsky, Alexandra S. Dubrovina, and Konstantin V. Kiselev. 2021. "The Influence of the Grapevine Bacterial and Fungal Endophytes on Biomass Accumulation and Stilbene Production by the In Vitro Cultivated Cells of Vitis amurensis Rupr." Plants 10, no. 7: 1276. https://doi.org/10.3390/plants10071276
APA StyleAleynova, O. A., Suprun, A. R., Nityagovsky, N. N., Dubrovina, A. S., & Kiselev, K. V. (2021). The Influence of the Grapevine Bacterial and Fungal Endophytes on Biomass Accumulation and Stilbene Production by the In Vitro Cultivated Cells of Vitis amurensis Rupr. Plants, 10(7), 1276. https://doi.org/10.3390/plants10071276