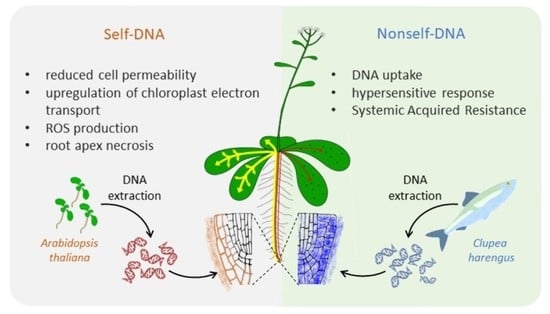
Arabidopsis thaliana Response to Extracellular DNA: Self Versus Nonself Exposure
Abstract
:1. Introduction
2. Results
2.1. Differential Gene Expression after Self and Nonself-DNA Exposure
2.2. GO Enrichment in Self and Nonself Treatments
2.3. Functional Response by Multivariate Analysis
2.4. Differentially Expressed Genes Associated to Enriched GOs
2.4.1. One Hour Post Treatment
2.4.2. Eight Hours Post Treatment
2.4.3. Sixteen Hours Post Treatment
2.5. Hormone Related DEGs after Self- and Nonself-DNA Treatments
2.6. DAMP and PAMP Associated Genes
2.7. QRT-PCR Results
2.8. Differential Self- and Nonself-DNA Distribution by Confocal Analysis and Phenotypic Changes in Seedlings
3. Discussion
3.1. Contrasting Transcriptome Dynamics in Response to Extracellular Self vs. Nonself-DNA
3.2. Hypotheses on the Mechanisms of Self-DNA Inhibition in Plants
3.3. EDAP: Extracellular DNA Associated Pathways
4. Materials and Methods
4.1. DNA Extraction and Preparation
4.2. Plant Materials and Treatments
4.3. Transcriptome and Bioinformatics Analyses
4.4. Confocal Microscopy Experiment
4.5. QRT-PCR Validation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mazzoleni, S.; Bonanomi, G.; Incerti, G.; Chiusano, M.L.; Termolino, P.; Mingo, A.; Senatore, M.; Giannino, F.; Cartenì, F.; Rietkerk, M.; et al. Inhibitory and toxic effects of extracellular self-DNA in litter: A mechanism for negative plant-soil feedbacks? New Phytol. 2015, 205, 1195–1210. [Google Scholar] [CrossRef] [Green Version]
- Mazzoleni, S.; Cartenì, F.; Bonanomi, G.; Senatore, M.; Termolino, P.; Giannino, F.; Incerti, G.; Rietkerk, M.; Lanzotti, V.; Chiusano, M.L. Inhibitory effects of extracellular self-DNA: A general biological process? New Phytol. 2015, 206, 127–132. [Google Scholar] [CrossRef]
- Mazzoleni, S.; Bonanomi, G.; Giannino, F.; Incerti, G.; Dekker, S.C.; Rietkerk, M. Modelling the effects of litter decomposition on tree diversity patterns. Ecol. Modell. 2010, 221, 2784–2792. [Google Scholar] [CrossRef]
- van der Putten, W.H.; Bradford, M.A.; Pernilla Brinkman, E.; van de Voorde, T.F.J.; Veen, G.F. Where, when and how plant–soil feedback matters in a changing world. Funct. Ecol. 2016, 30, 1109–1121. [Google Scholar] [CrossRef]
- Barbero, F.; Guglielmotto, M.; Capuzzo, A.; Maffei, M.E. Extracellular Self-DNA (esDNA), but Not Heterologous Plant or Insect DNA (etDNA), Induces Plasma Membrane Depolarization and Calcium Signaling in Lima Bean (Phaseolus lunatus) and Maize (Zea mays). Int. J. Mol. Sci. 2016, 17, 1659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, N.; Van der Putten, W.H.; Veen, G.F.C. Effects of root decomposition on plant-soil feedback of early- and mid-successional plant species. New Phytol. 2016, 212, 220–231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cartenì, F.; Bonanomi, G.; Giannino, F.; Incerti, G.; Vincenot, C.E.; Chiusano, M.L.; Mazzoleni, S. Self-DNA inhibitory effects: Underlying mechanisms and ecological implications. Plant Signal. Behav. 2016, 11, e1158381. [Google Scholar] [CrossRef] [Green Version]
- Veresoglou, S.D.; Aguilar-Trigueros, C.A.; Mansour, I.; Rillig, M.C. Self-DNA: A blessing in disguise? New Phytol. 2015, 207, 488–490. [Google Scholar] [CrossRef]
- Bhat, A.; Ryu, C.-M. Plant Perceptions of Extracellular DNA and RNA. Mol. Plant 2016, 9, 956–958. [Google Scholar] [CrossRef]
- Mazzoleni, S.; Cartenì, F.; Bonanomi, G.; Incerti, G.; Chiusano, M.L.; Termolino, P.; Migliozzi, A.; Senatore, M.; Giannino, F.; Rietkerk, M.; et al. New perspectives on the use of nucleic acids in pharmacological applications: Inhibitory action of extracellular self-DNA in biological systems. Phytochem. Rev. 2014, 13, 937–946. [Google Scholar] [CrossRef]
- Nagler, M.; Insam, H.; Pietramellara, G.; Ascher-Jenull, J. Extracellular DNA in natural environments: Features, relevance and applications. Appl. Microbiol. Biotechnol. 2018, 102, 6343–6356. [Google Scholar] [CrossRef] [Green Version]
- Pathan, S.I.; Arfaioli, P.; Ceccherini, M.T.; Ascher-Jenull, J.; Pietramellara, G. Preliminary evidences of the presence of extracellular DNA single stranded forms in soil. PLoS ONE 2020, 15, e0227296. [Google Scholar] [CrossRef]
- Collins, R.A.; Wangensteen, O.S.; O’Gorman, E.J.; Mariani, S.; Sims, D.W.; Genner, M.J. Persistence of environmental DNA in marine systems. Commun. Biol. 2018, 1, 185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agnelli, A.; Ascher, J.; Corti, G.; Ceccherini, M.T.; Pietramellara, G.; Nannipieri, P. Purification and isotopic signatures (δ 13C, δ 15N, Δ14C) of soil extracellular DNA. Biol. Fertil. Soils 2007, 44, 353–361. [Google Scholar] [CrossRef]
- Levy-Booth, D.J.; Campbell, R.G.; Gulden, R.H.; Hart, M.M.; Powell, J.R.; Klironomos, J.N.; Pauls, K.P.; Swanton, C.J.; Trevors, J.T.; Dunfield, K.E. Cycling of extracellular DNA in the soil environment. Soil Biol. Biochem. 2007, 39, 2977–2991. [Google Scholar] [CrossRef]
- Lejzerowicz, F.; Esling, P.; Majewski, W.; Szczuciński, W.; Decelle, J.; Obadia, C.; Arbizu, P.M.; Pawlowski, J. Ancient DNA complements microfossil record in deep-sea subsurface sediments. Biol. Lett. 2013, 9, 20130283. [Google Scholar] [CrossRef] [Green Version]
- Ascher, J.; Ceccherini, M.T.; Pantani, O.L.; Agnelli, A.; Borgogni, F.; Guerri, G.; Nannipieri, P.; Pietramellara, G. Sequential extraction and genetic fingerprinting of a forest soil metagenome. Appl. Soil Ecol. 2009, 42, 176–181. [Google Scholar] [CrossRef]
- Thomsen, P.F.; Willerslev, E. Environmental DNA—An emerging tool in conservation for monitoring past and present biodiversity. Biol. Conserv. 2015, 183, 4–18. [Google Scholar] [CrossRef]
- Hawes, M.C.; Curlango-Rivera, G.; Wen, F.; White, G.J.; Vanetten, H.D.; Xiong, Z. Extracellular DNA: The tip of root defenses? Plant Sci. 2011, 180, 741–745. [Google Scholar] [CrossRef] [PubMed]
- Wen, F.; Curlango-Rivera, G.; Huskey, D.A.; Xiong, Z.; Hawes, M.C. Visualization of extracellular DNA released during border cell separation from the root cap. Am. J. Bot. 2017, 104, 970–978. [Google Scholar] [CrossRef] [Green Version]
- Monticolo, F.; Palomba, E.; Termolino, P.; Chiaiese, P.; de Alteriis, E.; Mazzoleni, S.; Chiusano, M.L. The Role of DNA in the Extracellular Environment: A Focus on NETs, RETs and Biofilms. Front. Plant Sci. 2020, 11, 2045. [Google Scholar] [CrossRef]
- Paungfoo-Lonhienne, C.; Lonhienne, T.G.A.; Schmidt, S. DNA uptake by Arabidopsis induces changes in the expression of CLE peptides which control root morphology. Plant Signal. Behav. 2010, 5, 1112–1114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duran-Flores, D.; Heil, M. Growth inhibition by self-DNA: A phenomenon and its multiple explanations. New Phytol. 2015, 207, 482–485. [Google Scholar] [CrossRef] [PubMed]
- Panstruga, R. Bacterial RNA—A new MAMP on the block? New Phytol. 2016, 209, 458–460. [Google Scholar] [CrossRef] [Green Version]
- Vega-Muñoz, I.; Feregrino-Pérez, A.A.; Torres-Pacheco, I.; Guevara-González, R.G. Exogenous fragmented DNA acts as a damage-associated molecular pattern (DAMP) inducing changes in CpG DNA methylation and defence-related responses in Lactuca sativa. Funct. Plant Biol. 2018, 45, 1065–1072. [Google Scholar] [CrossRef] [PubMed]
- Heil, M.; Vega-Muñoz, I. Nucleic Acid Sensing in Mammals and Plants: Facts and Caveats. Int. Rev. Cell Mol. Biol. 2019, 345, 225–285. [Google Scholar] [CrossRef]
- Duran-Flores, D.; Heil, M. Extracellular self-DNA as a damage-associated molecular pattern (DAMP) that triggers self-specific immunity induction in plants. Brain Behav. Immun. 2018, 72, 78–88. [Google Scholar] [CrossRef] [PubMed]
- Toussaint, M.; Jackson, D.J.; Swieboda, D.; Guedán, A.; Tsourouktsoglou, T.-D.; Ching, Y.M.; Radermecker, C.; Makrinioti, H.; Aniscenko, J.; Bartlett, N.W.; et al. Host DNA released by NETosis promotes rhinovirus-induced type-2 allergic asthma exacerbation. Nat. Med. 2017, 23, 681–691. [Google Scholar] [CrossRef] [Green Version]
- Ren, X.; Chen, Z.; Liu, Y.; Zhang, H.; Zhang, M.; Liu, Q.; Hong, X.; Zhu, J.-K.; Gong, Z. ABO3, a WRKY transcription factor, mediates plant responses to abscisic acid and drought tolerance in Arabidopsis. Plant J. 2010, 63, 417–429. [Google Scholar] [CrossRef] [Green Version]
- Barrero, J.M.; Millar, A.A.; Griffiths, J.; Czechowski, T.; Scheible, W.R.; Udvardi, M.; Reid, J.B.; Ross, J.J.; Jacobsen, J.V.; Gubler, F. Gene expression profiling identifies two regulatory genes controlling dormancy and ABA sensitivity in Arabidopsis seeds. Plant J. 2010, 61, 611–622. [Google Scholar] [CrossRef]
- Santamaría, M.E.; Martínez, M.; Arnaiz, A.; Rioja, C.; Burow, M.; Grbic, V.; Díaz, I. An Arabidopsis TIR-Lectin Two-Domain Protein Confers Defense Properties against Tetranychus urticae. Plant Physiol. 2019, 179, 1298–1314. [Google Scholar] [CrossRef] [PubMed]
- Vanlerberghe, G.C.; McIntosh, L. Mitochondrial electron transport regulation of nuclear gene expression. Studies with the alternative oxidase gene of tobacco. Plant Physiol. 1994, 105, 867–874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dickman, M.; Williams, B.; Li, Y.; de Figueiredo, P.; Wolpert, T. Reassessing apoptosis in plants. Nat. Plants 2017, 3, 773–779. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.-H.; Kunz, H.-H.; Bhattacharjee, S.; Hauser, F.; Park, J.; Engineer, C.; Liu, A.; Ha, T.; Parker, J.E.; Gassmann, W.; et al. Natural Variation in Small Molecule-Induced TIR-NB-LRR Signaling Induces Root Growth Arrest via EDS1- and PAD4-Complexed R Protein VICTR in Arabidopsis. Plant Cell 2012, 24, 5177–5192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Enomoto, T.; Tokizawa, M.; Ito, H.; Iuchi, S.; Kobayashi, M.; Yamamoto, Y.Y.; Kobayashi, Y.; Koyama, H. STOP1 regulates the expression of HsfA2 and GDHs that are critical for low-oxygen tolerance in Arabidopsis. J. Exp. Bot. 2019, 70, 3297–3311. [Google Scholar] [CrossRef] [PubMed]
- Abd-El-Haliem, A.M.; Joosten, M.H.A.J. Plant phosphatidylinositol-specific phospholipase C at the center of plant innate immunity. J. Integr. Plant Biol. 2017, 59, 164–179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murray, S.L.; Ingle, R.A.; Petersen, L.N.; Denby, K.J. Basal resistance against Pseudomonas syringae in Arabidopsis involves WRKY53 and a protein with homology to a nematode resistance protein. Mol. Plant. Microbe Interact. 2007, 20, 1431–1438. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dutton, C.; Hõrak, H.; Hepworth, C.; Mitchell, A.; Ton, J.; Hunt, L.; Gray, J.E. Bacterial infection systemically suppresses stomatal density. Plant. Cell Environ. 2019, 42, 2411–2421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mandadi, K.K.; Misra, A.; Ren, S.; McKnight, T.D. BT2, a BTB Protein, Mediates Multiple Responses to Nutrients, Stresses, and Hormones in Arabidopsis. Plant Physiol. 2009, 150, 1930–1939. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dvořák, P.; Krasylenko, Y.; Ovečka, M.; Basheer, J.; Zapletalová, V.; Šamaj, J.; Takáč, T. FSD1: Developmentally-regulated plastidial, nuclear and cytoplasmic enzyme with anti-oxidative and osmoprotective role. Plant. Cell Environ. 2020. [Google Scholar] [CrossRef] [PubMed]
- Nishizawa, A.; Yabuta, Y.; Yoshida, E.; Maruta, T.; Yoshimura, K.; Shigeoka, S. Arabidopsis heat shock transcription factor A2 as a key regulator in response to several types of environmental stress. Plant J. 2006, 48, 535–547. [Google Scholar] [CrossRef]
- Cao, F.Y.; DeFalco, T.A.; Moeder, W.; Li, B.; Gong, Y.; Liu, X.-M.; Taniguchi, M.; Lumba, S.; Toh, S.; Shan, L.; et al. Arabidopsis ETHYLENE RESPONSE FACTOR 8 (ERF8) has dual functions in ABA signaling and immunity. BMC Plant Biol. 2018, 18, 211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coll, N.S.; Epple, P.; Dangl, J.L. Programmed cell death in the plant immune system. Cell Death Differ. 2011, 18, 1247–1256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belbin, F.E.; Noordally, Z.B.; Wetherill, S.J.; Atkins, K.A.; Franklin, K.A.; Dodd, A.N. Integration of light and circadian signals that regulate chloroplast transcription by a nuclear-encoded sigma factor. New Phytol. 2017, 213, 727–738. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Wang, Y.; Wang, L.; Liu, H.; Zhang, B.; Cao, Q.; Liu, X.; Lv, Y.; Bi, S.; Zhang, S.; et al. Arabidopsis PCaP2 Functions as a Linker Between ABA and SA Signals in Plant Water Deficit Tolerance. Front. Plant Sci. 2018, 9, 578. [Google Scholar] [CrossRef]
- Munemasa, S.; Hauser, F.; Park, J.; Waadt, R.; Brandt, B.; Schroeder, J.I. Mechanisms of abscisic acid-mediated control of stomatal aperture. Curr. Opin. Plant Biol. 2015, 28, 154–162. [Google Scholar] [CrossRef] [Green Version]
- Avalbaev, A.M.; Somov, K.A.; Yuldashev, R.A.; Shakirova, F.M. Cytokinin oxidase is key enzyme of cytokinin degradation. Biochemistry 2012, 77, 1354–1361. [Google Scholar] [CrossRef]
- Hou, S.; Liu, Z.; Shen, H.; Wu, D. Damage-Associated Molecular Pattern-Triggered Immunity in Plants. Front. Plant Sci. 2019, 10, 646. [Google Scholar] [CrossRef]
- Bacete, L.; Mélida, H.; Miedes, E.; Molina, A. Plant cell wall-mediated immunity: Cell wall changes trigger disease resistance responses. Plant J. 2018, 93, 614–636. [Google Scholar] [CrossRef]
- Li, Q.; Wang, C.; Mou, Z. Perception of Damaged Self in Plants. Plant Physiol. 2020, 182, 1545–1565. [Google Scholar] [CrossRef] [Green Version]
- Pham, A.Q.; Cho, S.-H.; Nguyen, C.T.; Stacey, G. Arabidopsis Lectin Receptor Kinase P2K2 Is a Second Plant Receptor for Extracellular ATP and Contributes to Innate Immunity. Plant Physiol. 2020, 183, 1364–1375. [Google Scholar] [CrossRef] [PubMed]
- Hornung, V.; Ablasser, A.; Charrel-Dennis, M.; Bauernfeind, F.; Horvath, G.; Caffrey, D.R.; Latz, E.; Fitzgerald, K.A. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature 2009, 458, 514–518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Greeff, C.; Roux, M.; Mundy, J.; Petersen, M. Receptor-like kinase complexes in plant innate immunity. Front. Plant Sci. 2012, 3, 209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferrusquía-Jiménez, N.I.; Chandrakasan, G.; Torres-Pacheco, I.; Rico-Garcia, E.; Feregrino-Perez, A.A.; Guevara-González, R.G. Extracellular DNA: A Relevant Plant Damage-Associated Molecular Pattern (DAMP) for Crop Protection Against Pests—A Review. J. Plant Growth Regul. 2020, 40, 451–463. [Google Scholar] [CrossRef]
- Albert, I.; Hua, C.; Nürnberger, T.; Pruitt, R.N.; Zhang, L. Surface Sensor Systems in Plant Immunity. Plant Physiol. 2020, 182, 1582–1596. [Google Scholar] [CrossRef] [Green Version]
- Cheung, A.Y.; Qu, L.-J.; Russinova, E.; Zhao, Y.; Zipfel, C. Update on Receptors and Signaling. Plant Physiol. 2020, 182, 1527–1530. [Google Scholar] [CrossRef] [Green Version]
- Gravino, M.; Locci, F.; Tundo, S.; Cervone, F.; Savatin, D.V.; De Lorenzo, G. Immune responses induced by oligogalacturonides are differentially affected by AvrPto and loss of BAK1/BKK1 and PEPR1/PEPR2. Mol. Plant Pathol. 2017, 18, 582–595. [Google Scholar] [CrossRef]
- De Lorenzo, G.; Ferrari, S.; Cervone, F.; Okun, E. Extracellular DAMPs in Plants and Mammals: Immunity, Tissue Damage and Repair. Trends Immunol. 2018, 39, 937–950. [Google Scholar] [CrossRef]
- Choi, H.W.; Klessig, D.F. DAMPs, MAMPs, and NAMPs in plant innate immunity. BMC Plant Biol. 2016, 16, 232. [Google Scholar] [CrossRef] [Green Version]
- Quintana-Rodriguez, E.; Duran-Flores, D.; Heil, M.; Camacho-Coronel, X. Damage-associated molecular patterns (DAMPs) as future plant vaccines that protect crops from pests. Sci. Hortic. 2018, 237, 207–220. [Google Scholar] [CrossRef]
- He, K.; Wu, Y. Receptor-Like Kinases and Regulation of Plant Innate Immunity. Enzymes 2016, 40, 105–142. [Google Scholar] [CrossRef]
- Bentham, A.R.; De la Concepcion, J.C.; Mukhi, N.; Zdrzałek, R.; Draeger, M.; Gorenkin, D.; Hughes, R.K.; Banfield, M.J. A molecular roadmap to the plant immune system. J. Biol. Chem. 2020, 295, 14916–14935. [Google Scholar] [CrossRef]
- Erb, M.; Reymond, P. Molecular Interactions Between Plants and Insect Herbivores. Annu. Rev. Plant Biol. 2019, 70, 527–557. [Google Scholar] [CrossRef] [Green Version]
- Bisgrove, S.R.; Simonich, M.T.; Smith, N.M.; Sattler, A.; Innes, R.W. A disease resistance gene in Arabidopsis with specificity for two different pathogen avirulence genes. Plant Cell 1994, 6, 927–933. [Google Scholar] [CrossRef] [Green Version]
- Clifton, R.; Millar, A.H.; Whelan, J. Alternative oxidases in Arabidopsis: A comparative analysis of differential expression in the gene family provides new insights into function of non-phosphorylating bypasses. Biochim. Biophys. Acta 2006, 1757, 730–741. [Google Scholar] [CrossRef] [Green Version]
- Selinski, J.; Hartmann, A.; Deckers-Hebestreit, G.; Day, D.A.; Whelan, J.; Scheibe, R. Alternative Oxidase Isoforms Are Differentially Activated by Tricarboxylic Acid Cycle Intermediates. Plant Physiol. 2018, 176, 1423–1432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maruta, T.; Tanouchi, A.; Tamoi, M.; Yabuta, Y.; Yoshimura, K.; Ishikawa, T.; Shigeoka, S. Arabidopsis Chloroplastic Ascorbate Peroxidase Isoenzymes Play a Dual Role in Photoprotection and Gene Regulation under Photooxidative Stress. Plant Cell Physiol. 2009, 51, 190–200. [Google Scholar] [CrossRef] [PubMed]
- Kliebenstein, D.J.; Monde, R.A.; Last, R.L. Superoxide dismutase in Arabidopsis: An eclectic enzyme family with disparate regulation and protein localization. Plant Physiol. 1998, 118, 637–650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bechtold, U.; Murphy, D.J.; Mullineaux, P.M. Arabidopsis Peptide Methionine Sulfoxide Reductase2 Prevents Cellular Oxidative Damage in Long Nights. Plant Cell 2004, 16, 908–919. [Google Scholar] [CrossRef] [Green Version]
- Simpson, P.J.; Tantitadapitak, C.; Reed, A.M.; Mather, O.C.; Bunce, C.M.; White, S.A.; Ride, J.P. Characterization of two novel aldo-keto reductases from Arabidopsis: Expression patterns, broad substrate specificity, and an open active-site structure suggest a role in toxicant metabolism following stress. J. Mol. Biol. 2009, 392, 465–480. [Google Scholar] [CrossRef]
- Seki, M.; Narusaka, M.; Ishida, J.; Nanjo, T.; Fujita, M.; Oono, Y.; Kamiya, A.; Nakajima, M.; Enju, A.; Sakurai, T.; et al. Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant J. 2002, 31, 279–292. [Google Scholar] [CrossRef] [PubMed]
- Werner, T.; Motyka, V.; Laucou, V.; Smets, R.; Van Onckelen, H.; Schmülling, T. Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 2003, 15, 2532–2550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.-Q.; Qu, X.-Q.; Hou, B.-H.; Sosso, D.; Osorio, S.; Fernie, A.R.; Frommer, W.B. Sucrose efflux mediated by SWEET proteins as a key step for phloem transport. Science 2012, 335, 207–211. [Google Scholar] [CrossRef]
- Eom, J.-S.; Chen, L.-Q.; Sosso, D.; Julius, B.T.; Lin, I.W.; Qu, X.-Q.; Braun, D.M.; Frommer, W.B. SWEETs, transporters for intracellular and intercellular sugar translocation. Curr. Opin. Plant Biol. 2015, 25, 53–62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanno, Y.; Oikawa, T.; Chiba, Y.; Ishimaru, Y.; Shimizu, T.; Sano, N.; Koshiba, T.; Kamiya, Y.; Ueda, M.; Seo, M. AtSWEET13 and AtSWEET14 regulate gibberellin-mediated physiological processes. Nat. Commun. 2016, 7, 13245. [Google Scholar] [CrossRef]
- Gallucci, S.; Maffei, M.E. DNA Sensing across the Tree of Life. Trends Immunol. 2017, 38, 719–732. [Google Scholar] [CrossRef] [PubMed]
- Heil, M.; Ibarra-Laclette, E.; Adame-Álvarez, R.M.; Martínez, O.; Ramirez-Chávez, E.; Molina-Torres, J.; Herrera-Estrella, L. How plants sense wounds: Damaged-self recognition is based on plant-derived elicitors and induces octadecanoid signaling. PLoS ONE 2012, 7, e30537. [Google Scholar] [CrossRef]
- Heil, M.; Land, W.G. Danger signals—Damaged-self recognition across the tree of life. Front. Plant Sci. 2014, 5, 578. [Google Scholar] [CrossRef]
- Duran-Flores, D.; Heil, M. Sources of specificity in plant damaged-self recognition. Curr. Opin. Plant Biol. 2016, 32, 77–87. [Google Scholar] [CrossRef]
- Wang, K.; Guo, Q.; Froehlich, J.E.; Hersh, H.L.; Zienkiewicz, A.; Howe, G.A.; Benning, C. Two Abscisic Acid-Responsive Plastid Lipase Genes Involved in Jasmonic Acid Biosynthesis in Arabidopsis thaliana. Plant Cell 2018, 30, 1006–1022. [Google Scholar] [CrossRef] [Green Version]
- Vos, I.; Verhage, A.; Schuurink, R.; Watt, L.; Pieterse, C.; Van Wees, S. Onset of herbivore-induced resistance in systemic tissue primed for jasmonate-dependent defenses is activated by abscisic acid. Front. Plant Sci. 2013, 4, 539. [Google Scholar] [CrossRef] [Green Version]
- Guo, Q.; Major, I.T.; Howe, G.A. Resolution of growth–defense conflict: Mechanistic insights from jasmonate signaling. Curr. Opin. Plant Biol. 2018, 44, 72–81. [Google Scholar] [CrossRef]
- Heil, M.; Baldwin, I.T. Fitness costs of induced resistance: Emerging experimental support for a slippery concept. Trends Plant Sci. 2002, 7, 61–67. [Google Scholar] [CrossRef]
- Pearce, G.; Yamaguchi, Y.; Barona, G.; Ryan, C.A. A subtilisin-like protein from soybean contains an embedded, cryptic signal that activates defense-related genes. Proc. Natl. Acad. Sci. USA 2010, 107, 14921–14925. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huffaker, A.; Pearce, G.; Veyrat, N.; Erb, M.; Turlings, T.C.J.; Sartor, R.; Shen, Z.; Briggs, S.P.; Vaughan, M.M.; Alborn, H.T.; et al. Plant elicitor peptides are conserved signals regulating direct and indirect antiherbivore defense. Proc. Natl. Acad. Sci. USA 2013, 110, 5707–5712. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, J.; Tanaka, K.; Cao, Y.; Qi, Y.; Qiu, J.; Liang, Y.; Lee, S.Y.; Stacey, G. Identification of a Plant Receptor for Extracellular ATP. Science 2014, 343, 290–294. [Google Scholar] [CrossRef]
- Klauser, D.; Desurmont, G.A.; Glauser, G.; Vallat, A.; Flury, P.; Boller, T.; Turlings, T.C.J.; Bartels, S. The Arabidopsis Pep-PEPR system is induced by herbivore feeding and contributes to JA-mediated plant defence against herbivory. J. Exp. Bot. 2015, 66, 5327–5336. [Google Scholar] [CrossRef] [Green Version]
- Poncini, L.; Wyrsch, I.; Dénervaud Tendon, V.; Vorley, T.; Boller, T.; Geldner, N.; Métraux, J.-P.; Lehmann, S. In roots of Arabidopsis thaliana, the damage-associated molecular pattern AtPep1 is a stronger elicitor of immune signalling than flg22 or the chitin heptamer. PLoS ONE 2017, 12, e0185808. [Google Scholar] [CrossRef] [Green Version]
- Jewell, J.B.; Sowders, J.M.; He, R.; Willis, M.A.; Gang, D.R.; Tanaka, K. Extracellular ATP Shapes a Defense-Related Transcriptome Both Independently and along with Other Defense Signaling Pathways. Plant Physiol. 2019, 179, 1144–1158. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.-W.; Jia, L.-Y.; Shi, D.-L.; Wang, R.; Lu, L.-N.; Xie, J.-J.; Sun, K.; Feng, H.-Q.; Li, X. Effects of extracellular ATP on local and systemic responses of bean (Phaseolus vulgaris L) leaves to wounding. Biosci. Biotechnol. Biochem. 2019, 83, 417–428. [Google Scholar] [CrossRef]
- Huang, H.-J.; Cui, J.-R.; Xia, X.; Chen, J.; Ye, Y.-X.; Zhang, C.-X.; Hong, X.-Y. Salivary DNase II from Laodelphax striatellus acts as an effector that suppresses plant defence. New Phytol. 2019, 224, 860–874. [Google Scholar] [CrossRef]
- Krol, E.; Mentzel, T.; Chinchilla, D.; Boller, T.; Felix, G.; Kemmerling, B.; Postel, S.; Arents, M.; Jeworutzki, E.; Al-Rasheid, K.A.S.; et al. Perception of the Arabidopsis danger signal peptide 1 involves the pattern recognition receptor AtPEPR1 and its close homologue AtPEPR2. J. Biol. Chem. 2010, 285, 13471–13479. [Google Scholar] [CrossRef] [Green Version]
- Lim, M.H.; Wu, J.; Clark, G.; Roux, S. Apyrases (NTPDases) and extracellular nucleotides regulate plant responses to biotic and abiotic stresses. Purinergic Signal. 2014, 10, 712. [Google Scholar]
- Yakushiji, S.; Ishiga, Y.; Inagaki, Y.; Toyoda, K.; Shiraishi, T.; Ichinose, Y. Bacterial DNA activates immunity in Arabidopsis thaliana. J. Gen. Plant Pathol. 2009, 75, 227–234. [Google Scholar] [CrossRef]
- Niehl, A.; Wyrsch, I.; Boller, T.; Heinlein, M. Double-stranded RNAs induce a pattern-triggered immune signaling pathway in plants. New Phytol. 2016, 211, 1008–1019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heidel, A.J.; Dong, X. Fitness Benefits of Systemic Acquired Resistance During Hyaloperonospora parasitica Infection in Arabidopsis thaliana. Genetics 2006, 173, 1621–1628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ecker, J.R.; Davis, R.W. Inhibition of gene expression in plant cells by expression of antisense RNA. Proc. Natl. Acad. Sci. USA 1986, 83, 5372–5376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hannon, G.J. RNA interference. Nature 2002, 418, 244–251. [Google Scholar] [CrossRef]
- Gruenert, D.; Bruscia, E.; Novelli, G.; Colosimo, A.; Dallapiccola, B.; Sangiuolo, F.; Goncz, K. Sequence-specific modification of genomic DNA by small DNA fragments. J. Clin. Invest. 2003, 112, 637–641. [Google Scholar] [CrossRef]
- Nisa, M.-U.; Huang, Y.; Benhamed, M.; Raynaud, C. The Plant DNA Damage Response: Signaling Pathways Leading to Growth Inhibition and Putative Role in Response to Stress Conditions. Front. Plant Sci. 2019, 10, 653. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.-H. Multifaceted Chromatin Structure and Transcription Changes in Plant Stress Response. Int. J. Mol. Sci. 2021, 22, 2013. [Google Scholar] [CrossRef]
- Luan, S. The CBL-CIPK network in plant calcium signaling. Trends Plant Sci. 2009, 14, 37–42. [Google Scholar] [CrossRef]
- Lee, K.-W.; Chen, P.-W.; Lu, C.-A.; Chen, S.; Ho, T.-H.D.; Yu, S.-M. Coordinated responses to oxygen and sugar deficiency allow rice seedlings to tolerate flooding. Sci. Signal. 2009, 2, ra61. [Google Scholar] [CrossRef] [PubMed]
- Pandey, G.K.; Kanwar, P.; Singh, A.; Steinhorst, L.; Pandey, A.; Yadav, A.K.; Tokas, I.; Sanyal, S.K.; Kim, B.-G.; Lee, S.-C.; et al. Calcineurin B-Like Protein-Interacting Protein Kinase CIPK21 Regulates Osmotic and Salt Stress Responses in Arabidopsis. Plant Physiol. 2015, 169, 780–792. [Google Scholar] [CrossRef] [Green Version]
- Cheng, N.-H.; Pittman, J.K.; Zhu, J.-K.; Hirschi, K.D. The protein kinase SOS2 activates the Arabidopsis H(+)/Ca(2+) antiporter CAX1 to integrate calcium transport and salt tolerance. J. Biol. Chem. 2004, 279, 2922–2926. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, W.-G.; Miller, G.; Wallace, I.; Harper, J.; Mittler, R.; Gilroy, S. Orchestrating rapid long-distance signaling in plants with Ca(2+), ROS and electrical signals. Plant J. 2017, 90, 698–707. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toyota, M.; Spencer, D.; Sawai-Toyota, S.; Jiaqi, W.; Zhang, T.; Koo, A.; Howe, G.; Gilroy, S. Glutamate triggers long-distance, calcium-based plant defense signaling. Science 2018, 361, 1112–1115. [Google Scholar] [CrossRef]
- Salvador-Recatalà, V.; Tjallingii, W.F.; Farmer, E.E. Real-time, in vivo intracellular recordings of caterpillar-induced depolarization waves in sieve elements using aphid electrodes. New Phytol. 2014, 203, 674–684. [Google Scholar] [CrossRef] [PubMed]
- Canales, J.; Henriquez-Valencia, C.; Brauchi, S. The Integration of Electrical Signals Originating in the Root of Vascular Plants. Front. Plant Sci. 2018, 8, 2173. [Google Scholar] [CrossRef] [Green Version]
- Chin, J.Y.; Glazer, P.M. Repair of DNA lesions associated with triplex-forming oligonucleotides. Mol. Carcinog. 2009, 48, 389–399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gahan, P.B.; Anker, P.; Stroun, M. Metabolic DNA as the origin of spontaneously released DNA? Ann. N. Y. Acad. Sci. 2008, 1137, 7–17. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [Green Version]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Howe, E.A.; Sinha, R.; Schlauch, D.; Quackenbush, J. RNA-Seq analysis in MeV. Bioinformatics 2011, 27, 3209–3210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef] [Green Version]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chiusano, M.L.; Incerti, G.; Colantuono, C.; Termolino, P.; Palomba, E.; Monticolo, F.; Benvenuto, G.; Foscari, A.; Esposito, A.; Marti, L.; et al. Arabidopsis thaliana Response to Extracellular DNA: Self Versus Nonself Exposure. Plants 2021, 10, 1744. https://doi.org/10.3390/plants10081744
Chiusano ML, Incerti G, Colantuono C, Termolino P, Palomba E, Monticolo F, Benvenuto G, Foscari A, Esposito A, Marti L, et al. Arabidopsis thaliana Response to Extracellular DNA: Self Versus Nonself Exposure. Plants. 2021; 10(8):1744. https://doi.org/10.3390/plants10081744
Chicago/Turabian StyleChiusano, Maria Luisa, Guido Incerti, Chiara Colantuono, Pasquale Termolino, Emanuela Palomba, Francesco Monticolo, Giovanna Benvenuto, Alessandro Foscari, Alfonso Esposito, Lucia Marti, and et al. 2021. "Arabidopsis thaliana Response to Extracellular DNA: Self Versus Nonself Exposure" Plants 10, no. 8: 1744. https://doi.org/10.3390/plants10081744
APA StyleChiusano, M. L., Incerti, G., Colantuono, C., Termolino, P., Palomba, E., Monticolo, F., Benvenuto, G., Foscari, A., Esposito, A., Marti, L., de Lorenzo, G., Vega-Muñoz, I., Heil, M., Carteni, F., Bonanomi, G., & Mazzoleni, S. (2021). Arabidopsis thaliana Response to Extracellular DNA: Self Versus Nonself Exposure. Plants, 10(8), 1744. https://doi.org/10.3390/plants10081744